Escolar Documentos
Profissional Documentos
Cultura Documentos
Enumeration
Enviado por
Sikin SikinTítulo original
Direitos autorais
Formatos disponíveis
Compartilhar este documento
Compartilhar ou incorporar documento
Você considera este documento útil?
Este conteúdo é inapropriado?
Denunciar este documentoDireitos autorais:
Formatos disponíveis
Enumeration
Enviado por
Sikin SikinDireitos autorais:
Formatos disponíveis
TITLE: ENUMERATION TECHNIQUE FOR VIABLE CELLS OBJECTIVE Counting the viable microorganisms by colony forming unit (CFU)
INTRODUCTION Counting bacteria is frequently important. For example, you may want to know the amount of bacteria in a sample such as the number of bacteria per millimetre of water in a swimming pool. Special techniques have been invented to enumerate bacteria, each with advantages and also disadvantages. There are three most common techniques namely plate count, direct count and turbidometric method. These counting of the number of colonies form are using colony forming unit (CFU) formula. Because it is not possible to be absolutely certain that each colony arose from an individual cell, the results are often referred or expressed in terms of colony forming unit (CFU) (Prescott L.M. et.al., 1999). Plate count is based on the premise that each viable bacterium produced a colony when growing on the agar plate. The material also known as the sample to be counted is suspended in a liquid and placed in an empty petri plate. The melted and cooled agar was poured to the plate and mixed with the inoculums. Each organisms produces a colony in the agar then can be counted after incubation period. The plate count is used very frequently but it has advantages and disadvantages. As for direct count, a suspension of bacteria is placed on the slide that has been ruled into squares and designed to hold a specific volume of liquid. By counting of bacteria that appear on the grid areas, the number of organisms can be calculated. Furthermore, this method is a little faster than plate count but it does have several drawbacks. Apart from these method, there is another method used that is turbidometric method. In this method of counting, a spectrophotometer is used to measure the turbidity or the optical density of bacteria in broth. The cloudier the broth means there are a lot of bacteria in it. Both direct and indirect counts are requiring that the organisms must uniformly disperse throughout the medium. If it is not uniformly dispersed, the result of the numbers of the count will not accurately represent the true microbial population in the entire sample. In some cases for example, the microorganisms were suspended in a liquid environment will commonly settle down to the bottom of the plate. In this case, such cultures must be resuspended again by swirling prior to the removal of a small volume for quantification. This isolation of pure culture method used can be spread plate and pour plate. Spread is an easy method to get a single colony by spread out the sample onto the surface of the agar medium. A small volume of a dilute microbial mixture is transferred to the centre of an agar plate and spread evenly with the L-shape rod (Prescott L.M et.al., 1999). For the 1
pour plate, the original sample is diluted several times to reduce the microbial population sufficiently to obtain separate colonies when plating. Then a small volume of several diluted samples are mixed with liquid agar that has been cooled and poured immediately into sterile dish. APPARATUS AND MATERIAL 1. Bunsen burner. 2. Universal bottle. 3. Agar plates. 4. L-shaped rod. 5. Micropipette METHOD SAMPLE PREPARATION 1. The amount of 10g (or ml) sample weighed on a bag stomacher and added 90ml saline solution to make a dilution factor of 10-1. 2. Sterilized pipette was used to transfer 1ml sample solution from 10-1 aseptically to other universal bottle containing 9ml saline solution. The serial dilution continued until it reaches the dilution factor of 10-6 and 10-7. A new pipette was used every new dilution was made. SPREAD PLATE TECHNIQUE 1. 0.1ml of the sample from the highest dilution factor was pipette onto the surface of the agar plate. The sample on the surface was then spread throughout the surface using the L-rod shaped. The L-rod shaped was first being sterilized by immersing it into ethanol a flame it into the fire. 2. The same step was repeated for the lower dilution. 3. The spread plate was then labelled and incubated for 24-36 hours. 4. After the period of incubation, the spread plate was observed and the colony formed was calculated. POUR PLATE TECHNIQUE 1. 1.0ml of sample from the highest dilution factor was transferred onto a sterilized empty Petri dish. The same step was done for the lower dilution factor. 2. Agar medium that was autoclaved and cooled was then poured aseptically onto the sample in the Petri dish until it covered all the bottom surface of the Petri dish. The 6. Ethanol. 7. Water bath. 8. Laminar air-flow. 9. Agar medium. 10. Food sample.
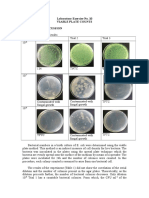
plate was then homogenized by making the 8 shape. it was then cooled down and solidified and incubated for 24-36 hours inverted. 3. After the period of incubation, do the calculation as usual. Result Figure 1: Spread Plate
From left to right: 10-2, 10-3
From left to right: 10-4, 10-5
Figure 2: Pour plate Pour plate
From left to right: 10-2, 10-3
From left to right: 10-4, 10-5
Table 1 the number of colony against the dilution factor for spread plate and pour plate Dilution factor 10-2 Spread plate Pour plate Average value colonies(pour plate and spread plate) UNC UNC 10-3 UNC UNC 10-4 202 97 10-5 149 52
Formula CFU value =
Since we only prepare one replicate for each plate dilution factor, therefore we did not average the colonies, 3
CFU value for spread plate at dilution factor of 10-4 = 2.02 x 107
CFU value for spread plate at dilution factor of 10-5 = 1.49 x 108
CFU value for pour plate at dilution factor of 10-4 = 9.7 x 105
CFU value for pour plate at dilution factor of 10-5 = 5.2 x 106
We also calculate the average colonies between pour plate and spread plate, CFU value for average colonies between spread plate and pour plate at dilution factor of 10-4 = 1.49 x 106
CFU value for average colonies between spread plate and pour plate at dilution factor of 10-5 = Discussion Enumeration of microorganism requires dilution of sample to achieve a population that is countable by the chosen method. Because it is difficult to count more than 3000 colonies on an agar plate, therefore it is necessary to dilute original bacteria culture before plate a known volume of the culture onto solid plate (Black, 2002). Generally, decimal or ten-fold dilutions are used for ease of calculation of final result. The serial dilution-agar plate (plate count) technique is based on the principle that when the material containing bacteria is cultured, every viable bacterium develops into a visible colony on a nutrient agar medium (Aneja, 2003). Based on result, the number of colonies in spread plate in dilution 10-2 and 10-3 is uncountable (UNC). However in spread plate of dilution 10-4, the number of colonies is 202, 4 1.0 x107
and in 10-5 is 149. For pour plate, the sample with dilution 10-2
and 10-3 both are
uncountable. Number of colonies in dilution factor dilution 10-4, the number of colonies is 97, and in 10-5 is 52. As for the calculation of CFU value, Since we only prepare one replicate for each plate dilution factor, therefore we did not average the colonies, For CFU value for spread plate at dilution factor of 10-4 is 2.02 x 107. While the CFU value for spread plate at dilution factor of 10-5 is 1.49 x 108. On the other hand CFU value for pour plate at dilution factor of 10-4 is 106. We also calculate the average colonies between pour plate and spread plate, CFU value for average colonies between spread plate and pour plate at dilution factor of 10-4 is 1.49 x 106 and CFU value for average colonies between spread plate and pour plate at dilution factor of 10-5 is 1.0 x107. For dilution factor of 10-2 1nd 10-3, the number of colonies are uncountable because the cell occur as clumps and stick as groups and it not easy to count is as a colonies. For the CFU value, it was calculated based on the number colonies that we observed and count under colony counter. However this is the estimation because mistake could happen during the experiment and the result might be not accurate as we think it is. From the result, we observed that the cell bacteria growing in aggregates. Some grow in clump or group form and some as single-cell. Therefore it difficult to counts the number of colonies. Therefore the there could be inaccuracy in determination of the colonies. The colony count methods provide an estimate number of viable microorganism in food according to the medium employed and the time and temperature of incubation. Microbial cell often occurs as clump or groups in food. Whereas shaking sample and dilution may uniformly distribute the clump of bacteria, this may not completely disrupt the clumps themselves. Mixing the initial dilution in a mechanical blender may provide better breakdown of clump. However, this does not ensure that the microorganism will be distributed as single cells. Consequently, each colony that appeared on the agar plates can arise from a clump of cells or from single cell and referred as colony forming unit. The failure of the microorganism to form visible colonies limits the accuracy of colony count method. This may result from deficiencies of nutrition, incubation temperature of unfavourable oxygen tension. In this experiment also, contamination may occur and cause the undesired microorganism to occur in agar. This can be cause by the dust circulation in 9.7 x 105 and CFU value for pour plate at dilution factor of 10-5 is 5.2 x
the lab or bacteria came from hand. Therefore it crucial to perform the experiment at laminar air flow to prevent contamination. There are advantages in using plate count techniques. The sensitivity of this technique is high and it allowed an inspection and positive identification of the organism counted. Plate count techniques is easy to perform and can be adapted to the measurement of populations of any magnitude. Since it is a sensitive technique, a small numbers or organisms can be counted. For instance, if a specimen contains a few as one bacterium per ml, one colony should develop up on the plating of 1 ml. However, plate count technique only living cells that develop colonies are counted. Clumps or chains of cells develop into a single colony. . A major disadvantage, however, is the time necessary for dilutions, platings and incubations, as well as the time needed for media preparation. Other limitation is colonies develop only from those organism for which the cultural condition are suitable for growth. Pour plate method plates are useful for quantifying microorganisms that grow in solid medium. It design to enumerate aerobic and facultative anaerobic bacteria in food, shellfish, water and dairy product (Messer et al., 1999). This method embeds colonies in agar. It is particularly useful for growing microaerophies that cannot tolerate exposure to oxygen in the air at the surface of the medium (Black, 2002). The disadvantage of this method is, the bacteria culture that pour with agar could heat-damaged, especially if the agar was pour when it still hot. This then result a smaller colonies that form inside the agar. In contrast with pour plate, instead of embedding microorganisms into agar, as is done with the pour plate method, in spread plate the liquid cultures are spread on the agar surface. An advantage of spreading a plate over the pour plate method is that cultures are never exposed to 45 oC (i.e. melted agar temperatures). Heat sensitive cells(ie. psychotroph) are not killed by the molten agar which may occur to an extent in the pour plate method. The spread plate eliminates the problem in pour plate method, because in spread plate case, the bacteria colonies will growth in surface of agar. This is result from the sample that spread on cooled agar surface evenly. The disadvantage of spread plate is there will be growth of more microbes and presence of more colony forming unit. In future experiment to get more accurate and reliable result, many of the error can be minimized. For instance, do a proper sterilization of diluents, media and equipment. Other than that, the sample measurement must accurate and the experiment must performed in
area that free from contamination. The sample should not be erratically mixing or shaking. Not only that, improper evaluation in counting and in computing count should not happen.
Você também pode gostar
- Lab SauerkrautDocumento6 páginasLab SauerkrautSikin Sikin100% (2)
- CLB11003 - Exp 4Documento6 páginasCLB11003 - Exp 4Nur DiyanahAinda não há avaliações
- Vapour Liquid Equilibrium ExpDocumento5 páginasVapour Liquid Equilibrium ExpAakash Sharma100% (1)
- Bsen 3310 Flow Behavior Lab Report 1Documento16 páginasBsen 3310 Flow Behavior Lab Report 1api-338795148Ainda não há avaliações
- Lab Report Reaction Use ThisDocumento19 páginasLab Report Reaction Use ThisKiran JojiAinda não há avaliações
- Lab Report Beer S LawDocumento16 páginasLab Report Beer S LawRhema Mohabul100% (2)
- A Financial Analysis of Alibaba Group Holding LTDDocumento26 páginasA Financial Analysis of Alibaba Group Holding LTDSrinu Gattu50% (4)
- Report BacteriaDocumento11 páginasReport BacteriaSuzeanni JalilAinda não há avaliações
- Serial DilutionDocumento9 páginasSerial DilutionSaranya KannanAinda não há avaliações
- Serial Dilution MethodDocumento3 páginasSerial Dilution Methoddraneesh75% (4)
- Jordi PD LabDocumento3 páginasJordi PD LabAaron Li100% (1)
- CHM170L - Final Report 3Documento5 páginasCHM170L - Final Report 3Katrina BucudAinda não há avaliações
- Biology Lab Report AntibioticsDocumento8 páginasBiology Lab Report AntibioticsLoore Liflander100% (1)
- BIOL 240 Lab Report 1Documento11 páginasBIOL 240 Lab Report 1Ben CharlesAinda não há avaliações
- Lab Report 1 Diffusion Across Biological Membrances SimulationDocumento7 páginasLab Report 1 Diffusion Across Biological Membrances SimulationAmeena Ali50% (2)
- Experiment 5 Determination of Caffeine 2020Documento4 páginasExperiment 5 Determination of Caffeine 2020FYAinda não há avaliações
- Lab YogurtDocumento6 páginasLab YogurtSikin Sikin0% (1)
- Serial Dilution MethodDocumento3 páginasSerial Dilution Methoddraneesh75% (4)
- Lab Preservation 2Documento6 páginasLab Preservation 2Sikin SikinAinda não há avaliações
- CibaDocumento16 páginasCibamaheshgupte0% (1)
- Serial Dilution ProtocolsDocumento5 páginasSerial Dilution ProtocolsYugendra Babu K100% (1)
- Laboratory Exercise No. 10 Viable Plate Counts Results and DiscussionDocumento3 páginasLaboratory Exercise No. 10 Viable Plate Counts Results and Discussionvanessa olga100% (1)
- Lab # 7 - Serial DilutionDocumento2 páginasLab # 7 - Serial DilutionKESHAWN TYRONE SUTHERLANDAinda não há avaliações
- Lab Report Enumeration of BacteriaDocumento13 páginasLab Report Enumeration of BacteriaA'ziemullah UmarAinda não há avaliações
- Microbiology-Microbial GrowthDocumento8 páginasMicrobiology-Microbial GrowthViAn100% (2)
- Lab Report For Micro 210 AJH1Documento11 páginasLab Report For Micro 210 AJH1ahathaw17Ainda não há avaliações
- Streak PlatesDocumento0 páginaStreak PlatesJenry Reyes TrispaAinda não há avaliações
- ELA Heat of SolutionDocumento15 páginasELA Heat of SolutionJimAinda não há avaliações
- Microbiology Lab Report 3Documento5 páginasMicrobiology Lab Report 3FarhinAinda não há avaliações
- Report Case Study FluidDocumento11 páginasReport Case Study FluidAnisalya JamriAinda não há avaliações
- CSTR in SeriesDocumento17 páginasCSTR in SeriesDhiyyah MardhiyyahAinda não há avaliações
- Heat Solution LabDocumento6 páginasHeat Solution LabNicole Graham50% (2)
- Bubble Cap Distillation ColumnDocumento3 páginasBubble Cap Distillation Columnnhalieza1067Ainda não há avaliações
- Practical Manual of Extrusion Technology FST-608 (Dr. Faiz)Documento16 páginasPractical Manual of Extrusion Technology FST-608 (Dr. Faiz)Ayesha SafdarAinda não há avaliações
- Experiment: Packed Distillation ColumnDocumento4 páginasExperiment: Packed Distillation Columnnhalieza1067Ainda não há avaliações
- Reaction Rate and Rate Constant of The Hydrolysis of Ethyl Acetate With Sodium HydroxideDocumento5 páginasReaction Rate and Rate Constant of The Hydrolysis of Ethyl Acetate With Sodium HydroxideAmyAinda não há avaliações
- Fractional DistillationDocumento5 páginasFractional Distillationihack_101Ainda não há avaliações
- PDFDocumento88 páginasPDFMuralidharanAinda não há avaliações
- Detection and Enumeration of The Most Probable Number of ColiformsDocumento6 páginasDetection and Enumeration of The Most Probable Number of ColiformsLaksilu Viduraga Peiris83% (6)
- Suspension Od Solid Particles (Revised Report)Documento7 páginasSuspension Od Solid Particles (Revised Report)michsantosAinda não há avaliações
- Tray DryerDocumento16 páginasTray Dryermirdza94Ainda não há avaliações
- Exp. 8 Diffusion of Sodium Chloride in WaterDocumento6 páginasExp. 8 Diffusion of Sodium Chloride in WaterElaine Pui100% (1)
- Recombinant ReportDocumento4 páginasRecombinant Report门门Ainda não há avaliações
- Determination of Concentration of ChromiumDocumento26 páginasDetermination of Concentration of ChromiumCik Tiem Ngagiman70% (10)
- Lab Report - Distillation of Bubble CapDocumento21 páginasLab Report - Distillation of Bubble Capratish100% (1)
- Experiment 1 Introduction and ConclusionDocumento3 páginasExperiment 1 Introduction and ConclusionMaiSakurajima100% (1)
- Estimation of Dissolved Carbon DioxideDocumento2 páginasEstimation of Dissolved Carbon DioxideJR ParkAinda não há avaliações
- Q2015 Physical Chemistry Measurements Laboratory Chemistry Department, Campus Monterrey Practice # 8 Distribution CoefficientDocumento4 páginasQ2015 Physical Chemistry Measurements Laboratory Chemistry Department, Campus Monterrey Practice # 8 Distribution Coefficientandres_guadiana_7362100% (1)
- Effect of PH On Enzyme Activity Lab 3Documento8 páginasEffect of PH On Enzyme Activity Lab 3api-340907023Ainda não há avaliações
- Experiment 3Documento14 páginasExperiment 3HafiniHambaliAinda não há avaliações
- SedimentationDocumento7 páginasSedimentationgrkhari1100% (2)
- Climacteric Fruit Ripening PDFDocumento7 páginasClimacteric Fruit Ripening PDFsandraosouzaAinda não há avaliações
- Osbourne ReynoldDocumento13 páginasOsbourne ReynoldN Afiqah Razak0% (1)
- Lab Report Experiment 1Documento10 páginasLab Report Experiment 1Arieanna AsyiqinAinda não há avaliações
- Lab Report CHM260 (Exp 2)Documento19 páginasLab Report CHM260 (Exp 2)Nur AthirahAinda não há avaliações
- Microbial Growth and Product FormationDocumento26 páginasMicrobial Growth and Product FormationDP PurwadiAinda não há avaliações
- ) Is 0.6. The Feed Stream Is SterileDocumento3 páginas) Is 0.6. The Feed Stream Is SterileAA0809Ainda não há avaliações
- Exp 4 Batch Evaporative Crystallization PDFDocumento9 páginasExp 4 Batch Evaporative Crystallization PDFmirza farhanAinda não há avaliações
- Lab Report To Determine The Concentration Using GC-MSDocumento9 páginasLab Report To Determine The Concentration Using GC-MSSamuel Ogeda OtienoAinda não há avaliações
- Diffusion Coefficient Full Report TiqaDocumento19 páginasDiffusion Coefficient Full Report TiqaprmzAinda não há avaliações
- 5 - (CSTR Bp143)Documento12 páginas5 - (CSTR Bp143)Aisyah Addia AzizanAinda não há avaliações
- NITHYA Micro Exp3Documento9 páginasNITHYA Micro Exp3Nithyakalyani AsokanAinda não há avaliações
- Experiment 5Documento5 páginasExperiment 5Nipuni Saubhagya RathnayakeAinda não há avaliações
- Bacterial TitreDocumento3 páginasBacterial TitreManjunatha Bukkambudi KrishnaswamyAinda não há avaliações
- UntitledDocumento30 páginasUntitledLEE JIA XINAinda não há avaliações
- Experiment - Viable Count PDFDocumento15 páginasExperiment - Viable Count PDFImzafreenAinda não há avaliações
- Ishikawa DiagramDocumento1 páginaIshikawa DiagramSikin SikinAinda não há avaliações
- TT 0309Documento2 páginasTT 0309Sikin SikinAinda não há avaliações
- SpectroscopDocumento104 páginasSpectroscopSikin SikinAinda não há avaliações
- Ishikawa DiagramDocumento1 páginaIshikawa DiagramSikin SikinAinda não há avaliações
- (Your Name & Full Address) (Date) : Li Placement Application Cover Letter SampleDocumento2 páginas(Your Name & Full Address) (Date) : Li Placement Application Cover Letter SampleSikin SikinAinda não há avaliações
- (Company's Name & Full Address) : UMS/SSMP6.8/600-3/15/1 (Date)Documento1 página(Company's Name & Full Address) : UMS/SSMP6.8/600-3/15/1 (Date)Sikin SikinAinda não há avaliações
- Universiti Malaysia Sabah: Senarai Semak Latihan IndustriDocumento3 páginasUniversiti Malaysia Sabah: Senarai Semak Latihan IndustriSikin SikinAinda não há avaliações
- Practicum Report - Doc081111Documento2 páginasPracticum Report - Doc081111Sikin SikinAinda não há avaliações
- 9th LectureDocumento5 páginas9th LectureSikin SikinAinda não há avaliações
- RUJUKANDocumento2 páginasRUJUKANSikin SikinAinda não há avaliações
- Ii 1213Documento5 páginasIi 1213Sikin SikinAinda não há avaliações
- Syllabus CorpEntre 2012Documento7 páginasSyllabus CorpEntre 2012Sikin SikinAinda não há avaliações
- TT Dealer - East MsiaDocumento4 páginasTT Dealer - East MsiaSikin SikinAinda não há avaliações
- Practical Training & Practical Report Writing: GuidebookDocumento14 páginasPractical Training & Practical Report Writing: Guidebookawg_ridhwanAinda não há avaliações
- Microbiology Lab 3Documento6 páginasMicrobiology Lab 3Sikin SikinAinda não há avaliações
- Preservation of Vegetables by Salting or BriningDocumento9 páginasPreservation of Vegetables by Salting or BriningagungpuruhitaAinda não há avaliações
- Lab Awet 5Documento3 páginasLab Awet 5Sikin SikinAinda não há avaliações
- 8th LectureDocumento9 páginas8th LectureSikin SikinAinda não há avaliações
- 3rd LectureDocumento4 páginas3rd LectureSikin SikinAinda não há avaliações
- Eastern and Western Cookery SinopsisDocumento9 páginasEastern and Western Cookery SinopsisSikin SikinAinda não há avaliações
- Lab Preservation 1Documento7 páginasLab Preservation 1Sikin SikinAinda não há avaliações
- Fud Chem ResultDocumento1 páginaFud Chem ResultSikin SikinAinda não há avaliações
- Spray Dryer PRNT SCRNDocumento2 páginasSpray Dryer PRNT SCRNSikin SikinAinda não há avaliações
- Instant Download Developing Management Skills 10th Edition Whetten Test Bank PDF Full ChapterDocumento33 páginasInstant Download Developing Management Skills 10th Edition Whetten Test Bank PDF Full Chapterdonaldvioleti7o100% (9)
- Daftar PustakaDocumento2 páginasDaftar PustakaBang UsopAinda não há avaliações
- Q400 PropellerDocumento10 páginasQ400 PropellerMoshiurRahman100% (1)
- Microsoft Lumia 950 XL - Unlocked (Black)Documento8 páginasMicrosoft Lumia 950 XL - Unlocked (Black)Dawood AhmedAinda não há avaliações
- International Business of Pizza HutDocumento13 páginasInternational Business of Pizza Hutpratikdotia9100% (2)
- Am No. 98-5-01-SC, Nov 9 1998 in Re Hon Mateo ValenzuelaDocumento9 páginasAm No. 98-5-01-SC, Nov 9 1998 in Re Hon Mateo Valenzuelaian clark MarinduqueAinda não há avaliações
- 01 B744 B1 ATA 23 Part2Documento170 páginas01 B744 B1 ATA 23 Part2NadirAinda não há avaliações
- 2016 May Virginia Medical Law ReportDocumento20 páginas2016 May Virginia Medical Law ReportMichael DuntzAinda não há avaliações
- People vs. Samson, September 2, 2015Documento5 páginasPeople vs. Samson, September 2, 2015Alpha Grace JugalAinda não há avaliações
- Scottish Myths and Legends (PDFDrive)Documento181 páginasScottish Myths and Legends (PDFDrive)VeraAinda não há avaliações
- Article 21Documento17 páginasArticle 21shanikaAinda não há avaliações
- The 5 Best 5G Use Cases: Brian SantoDocumento4 páginasThe 5 Best 5G Use Cases: Brian SantoabdulqaderAinda não há avaliações
- Cold SandwichesDocumento7 páginasCold SandwichesLea Galano ManarpaacAinda não há avaliações
- Statement by The Ukrainian Canadian CongressDocumento1 páginaStatement by The Ukrainian Canadian CongressLevon SevuntsAinda não há avaliações
- Chapter Three: The Topography of Ethiopia and The HornDocumento4 páginasChapter Three: The Topography of Ethiopia and The Horneyob astatke100% (1)
- My Slow Carb Diet Experience, Hacking With Four Hour BodyDocumento37 páginasMy Slow Carb Diet Experience, Hacking With Four Hour BodyJason A. Nunnelley100% (2)
- Global Value Chain: Shikha GuptaDocumento19 páginasGlobal Value Chain: Shikha GuptaRushilAinda não há avaliações
- Seven Days in The Art World - Sarah ThorntonDocumento249 páginasSeven Days in The Art World - Sarah ThorntonTô Tiêu NgọcAinda não há avaliações
- Orange PeelDocumento2 páginasOrange PeelCharul Shukla100% (1)
- Nunez Rodarte v. Holder, JR., 10th Cir. (2010)Documento7 páginasNunez Rodarte v. Holder, JR., 10th Cir. (2010)Scribd Government DocsAinda não há avaliações
- Disediakan Oleh: Keetha Naganthiran: Set Induction (Starter) Main ActivitiesDocumento1 páginaDisediakan Oleh: Keetha Naganthiran: Set Induction (Starter) Main ActivitiesnandyshaAinda não há avaliações
- Cristoforo BuondelmontiDocumento15 páginasCristoforo BuondelmontiAnna AchiolaAinda não há avaliações
- Hampers 2023 - Updted Back Cover - FADocumento20 páginasHampers 2023 - Updted Back Cover - FAHaris HaryadiAinda não há avaliações
- 2463-Article Text-6731-1-10-20220712Documento18 páginas2463-Article Text-6731-1-10-20220712Okto LedohAinda não há avaliações
- BHCS31352F Mentor ViQ Specsheet - R24Documento8 páginasBHCS31352F Mentor ViQ Specsheet - R24Jocélio A. TavaresAinda não há avaliações
- Physical and Chemical Changes WorksheetDocumento2 páginasPhysical and Chemical Changes Worksheetisabe;llaAinda não há avaliações
- Safari - 15 Nov 2023 at 8:34 PMDocumento1 páginaSafari - 15 Nov 2023 at 8:34 PMrgk966c275Ainda não há avaliações
- Structuralism From Bsa 2D PDFDocumento57 páginasStructuralism From Bsa 2D PDFPrincess AlegreAinda não há avaliações