Escolar Documentos
Profissional Documentos
Cultura Documentos
Ubiquinone Synthesis
Enviado por
Jesus LirioDescrição original:
Direitos autorais
Formatos disponíveis
Compartilhar este documento
Compartilhar ou incorporar documento
Você considera este documento útil?
Este conteúdo é inapropriado?
Denunciar este documentoDireitos autorais:
Formatos disponíveis
Ubiquinone Synthesis
Enviado por
Jesus LirioDireitos autorais:
Formatos disponíveis
Vol. 47 No.
2/2000
469480 QUARTERLY
Review
Ubiquinone. Biosynthesis of quinone ring and its isoprenoid side chain. Intracellular localization.
Anna Szkopiska
Department of Lipid Biochemistry, Institute of Biochemistry and Biophysics, Polish Academy of Sciences, A. Pawiskiego 5a, 02-106 Warszawa, Poland
Received: 19 January, 2000; accepted: 17 May, 2000
Key words: ubiquinone, polyisoprenoid diphosphates, polyprenyltransferases Ubiquinone, known as coenzyme Q, was shown to be the part of the metabolic pathways by Crane et al. in 1957. Its function as a component of the mitochondrial respiratory chain is well established. However, ubiquinone has recently attracted increasing attention with regard to its function, in the reduced form, as an antioxidant. In ubiquinone synthesis the para-hydroxybenzoate ring (which is the derivative of tyrosine or phenylalanine) is condensed with a hydrophobic polyisoprenoid side chain, whose length varies from 6 to 10 isoprene units depending on the organism. para-Hydroxybenzoate (PHB) polyprenyltransferase that catalyzes the condensation of PHB with polyprenyl diphosphate has a broad substrate specificity. Most of the genes encoding (all-E)-prenyltransferases which synthesize polyisoprenoid chains, have been cloned. Their structure is either homo- or heterodimeric. Genes that encode prenyltransferases catalysing the transfer of the isoprenoid chain to para-hydroxybenzoate were also cloned in bacteria and yeast. To form ubiquinone, prenylated PHB undergoes several modifications such as hydroxylations, O-methylations, methylations and decarboxylation. In eukaryotes ubiquinones were found in the inner mitochondrial membrane and in other membranes such as the endoplasmic reticulum, Golgi vesicles, lysosomes and peroxisomes. Still, the subcellular site of their biosynthesis remains unclear. Considering the diversity of functions of ubiquinones, and their multistep biosynthesis, identification of factors regulating their cellular level remains an elusive task.
This work was supported by Grant 6 PO4A 020 13 from the State Committee for Scientific Research and by the French-Polish Center of Plant Biotechnology. To whom correspondence should be addressed; fax: (48 22) 3912 1623; e-mail: babel@ibbrain.ibb.waw.pl Abbreviations: PHB, para-hydroxybenzoate (4-hydroxybenzoate); HMG-CoA, 3-hydroxy-3-methylglutaryl coenzyme A; IPP, isopentenyl diphosphate; DMAPP, dimethylallyl diphosphate, GPP, geranyl diphosphate; FPP, farnesyl diphosphate; GGPP, geranylgeranyl diphosphate; HexPP, hexaprenyl diphosphate; HepPP, heptaprenyl diphosphate.
470
A. Szkopiska
2000
Ubiquinone is a component of the mitochondrial respiratory chain [1], participating in electron transport in NADH-coenzyme Q reductase (complex I), succinate coenzyme Q reductase (complex II) and the cytochrome system. Folkers and his group [2] determined the structure of the quinone moiety which was found identical to that described by Morton and his team [3], and suggested the name ubiquinone referring to the ubiquitous occurrence of this compound in various tissues. The name ubiquinone (coenzyme Q) was officially accepted in 1975 by the IUPAC-IUB Commission on Biochemical Nomenclature. The growing interest in ubiquinones is fully justified. In addition to participating in the respiratory chain, they are involved in the redox processes taking place in cytoplasmic as well as Golgi system membranes. In reduced form, as antioxidants, they efficiently protect membrane phospholipids and lipoproteins from lipid peroxidation, as well as membrane proteins and DNA from free radical-induced oxidative damage [46]. In plants similar roles are played by plastoquinones [7]. Some vitamins (E, K) are quinone derivatives. The development of molecular biology and recombinant DNA technologies, the discovery of restriction enzymes and construction of vectors made new approaches possible to investigate ubiquinones. They allowed the identification and cloning of genes encoding enzymes participating in modifications of the ring moiety of the ubiquinone molecule; methyltransferases: UBIG, UBIE from bacteria and COQ3 from yeast; hydroxylases UBIB, UBIH from bacteria and COQ6 from yeast. Genes encoding prenyltransferases catalyzing the transfer of the isoprenoid chain to para-hydroxybenzoate in bacteria (UBIA) and yeast (COQ2) have also been cloned. Practically all the bacterial genes responsible for the synthesis of hexa- up to decaprenyl diphosphate synthases forming the ubiquinone polyisoprenoid side chain have been cloned. These results will allow identification of the respective human genes, characterization of their
structure, level and site of expression which is of vital importance considering the functions of ubiquinones.
SYNTHESIS OF QUINONE MOIETY
The benzene moiety is derived mainly from tyrosine (in some cases from phenylalanine) converted to para-hydroxybenzoate [8, 9] (Fig. 1) which in turn is condensed with all-E polyisoprenoid diphosphate. A number of subsequent modifications of the ring are required for the completion of ubiquinone [10] (Fig. 2). Modifications of para-hydroxybenzoate (4-OH-benzoate) condensed with a polyisoprenoid side chain start with C-hydroxylation, followed by O-methylation and decarboxylation. Two additional C-hydroxylations and one O-methylation are necessary for the final synthesis of ubiquinone. The first data on methylation and O-methylation of the benzene ring come from the elegant chemical work of Olson et al. [11]. The above sequence of events has been established in a bacterial system [12]. However, the results obtained by Kang et al. [13] suggest that in an animal system decarboxylation may occur prior to the first methylation [13]. Genes UBIG and COQ3 encode an S-adenosylmethionine O-methyl transferase involved in O-methylation in bacteria and yeast, respectively (Fig. 2) [14, 15]. The UBIH gene encodes a mono-oxygenase thought to contain flavin adenine nucleotide that catalyses the conversion of the 6-octaprenyl-2-methoxyphenol to 6-octaprenyl2-methoxy-1,4-benzoquinone [16]. Recently a novel gene ohb1 encoding a reversible parahydroxybenzoate decarboxylase from Clostridium hydroxybenzoicum was cloned and characterized [17]. Its amino-acid sequence shows 57% identity and 74% similarity to that of hypothetical proteins deduced from open reading frames in genomes from bacteria and archaea, suggesting the possible existence of a novel gene family. These genes could have encoded an ancient type of decarboxylase, which
Vol. 47
Ubiquinone biosynthesis
471
Figure 1. Multistep biosynthesis of para-hydroxybenzoate. 1, Phenylalanine or 2, tyrosine amino-acid precursors; 3, para-hydroxyphenylpyruvate; 4, para-hydroxyphenyllactate; 5, para-hydroxycinnamate; 6, para-hydroxybenzoate (4-hydroxybenzoate) [50].
was replaced during evolution by a more efficient one [17]. Not all the genes that encode enzymes participating in ubiquinone biosynthesis in higher eukaryotic cells have been cloned. It seems however, that they are similar to the yeast genes. Indeed, a rat cDNA homologue to UBIG and COQ3 has been cloned by functional complementation of a yeast coq3 mutant with a rat testis cDNA library [18]. In plants the biosynthesis of ubiquinones starting from isopentenyl diphosphate and para-
the proteins nor the corresponding genes have been characterized so far [19].
SYNTHESIS OF THE POLYPRENYL SIDE CHAIN
The polyprenyl (isoprenoid) side chain of ubiquinone is synthesized from acetyl-CoA through a sequence of reaction named the mevalonic acid pathway, leading to the formation of farnesyl diphosphate (FPP) (Fig. 3).
Figure 2. The pathway of ubiquinone biosynthesis from 4-hydroxybenzoate [76]. PPHB, polyprenyl para-hydroxybenzoate; COQ2, Saccharomyces cerevisiae gene encoding prenyltransferase catalysing the transfer of the polyisoprenoid chain to para-hydroxybenzoate; COQ3, S. cerevisiae gene encoding an S-adenosylmethionine O-methyl transferase.
hydroxybenzoate also requires a similar set of reactions i.e. hydroxylation, decarboxylation, O-methylation and methylation but neither
The study of the isoprenoid biosynthesis was brought to an enzymatical level by the discovery made independently by the groups of
472
A. Szkopiska
2000
Figure 3. The mevalonic acid pathway.
Lynen and Bloch in 1958 [20, 21] of isopentenyl diphosphate (IPP) as the active isoprene unit. It is worth mentioning that Rohmer et al. [22] discovered a novel biosynthesis pathway of IPP in which mevalonate is not involved. On the basis of labeling patterns of bacterial hopanoids and ubiquinones derived from 13 C-labeled metabolites of glycolysis as well as from [13C]acetate, they proved that glyceraldehyde 3-phosphate occupies the branch point of the glycolysis leading to the non-mevalonate pathway synthesizing IPP [22, 23]. The discovery of IPP isomerase, which converts IPP to dimethylallyl diphosphate (DMAPP) [24], as well as the prenyltransfe-
rase that catalyses the head-to-tail condensations of IPP with DMAPP and with geranyl diphosphate (GPP), to form (E,E)-farnesyl diphosphate (FPP) (Fig. 4) has led to the recognition that a tremendous number of isoprenoid compounds exists in Nature. The chain length of prenyl diphosphates varies ranging from geraniol (C10) to natural rubber whose carbon chain length extends to several millions. To date 16 prenyltransferases (enzymes that catalyze the transfer of prenyl groups to an acceptor which is usually IPP, but may also be an aromatic compound, a protein etc) with different catalytic functions have been characterized. In contrast to dolichols composed of 14 to 23 isoprene units in yeast and man, the length of the side chain of ubiquinone varies to a much more limited extent. For instance, Escherichia coli produces mainly ubiquinones having 8 isoprene units, Saccharomyces cerevisiae, 6 isoprene units. Eukaryotic cells contain ubiquinones with 610 isoprene residues but in every species only one chain length dominates. Organisms constitutively contain at least one of the short-chain prenyl diphosphate synthases FPP (geranyltranstransferase, EC 2.5.1.10) or GGPP (farnesyltranstransferase, EC 2.5.1.30) for the production of prenyl diphosphates which act as the priming substrates for other groups of prenyltransferases. The short-chain prenyl diphosphates are also biosynthetic precursors of various isoprenoids including steroids, carotenoids and prenylated proteins. It is noteworthy that
Figure 4. The two-step reaction catalyzed by farnesyl diphosphate (FPP) synthase. IPP, isopentenyl diphosphate; DMAPP, dimethylallyl diphosphate; GPP, geranyl diphosphate.
Vol. 47
Ubiquinone biosynthesis
473
GGPP (geranylgeranyl diphosphate) synthase occurs not only in bacteria and plants but also in mammals. FPP (Fig. 5) and GGPP synthases both have a homodimeric structure in which the subunits are tightly bound to each other [25]. In spite of the fact that all eukaryotic cells probably contain E-prenyltransferase activity, to date only a few investigations have been concerned with the characterization of this
which occurs in all bacteria. HexPP synthase has been cloned from Micrococcus luteus B-P 26, and sequenced [28]. Within a 2130 bp DNA which expresses HexPP synthase in E. coli cells, there are three consecutive open reading frames (ORF1 143 aa, ORF2 246 aa and ORF3 325 aa). It has been established that the two dissociable components A and B of HexPP synthase are products of ORF1 and ORF3, respectively1. These genes
Figure 5. Yeast farnesyl diphosphate synthase homodimer model. The model was constructed according to the avian crystal structure [25] by D. Pochocka from Bioinformatic Dept. of Inst. Biochem. Biophys. PAS, Warszawa. One monomer is in ribbon representation, the other is shown as a backbone.
enzyme in animal cells. On the other hand, bacteria are good sources of prenyltransferases because they produce ubiquinones and menaquinones with polyprenyl side chains of chain lengths varying in a species-specific manner [26].
Hexaprenyl diphosphate synthases
It has been found that the quinone side chain consisting of 6 isoprene units is synthesized by hexaprenyl diphosphate (HexPP) synthase [EC 2.5.1.30] which has a novel subunit system (Fig. 6). The enzyme catalyzes the synthesis of (all-E)-HexPP by adding three molecules of IPP to FPP, but it can not catalyze the steps of synthesis of GPP or FPP from DMAPP and IPP [27], FPP is supplied by FPP synthase
1
were similarly located in a newly identified gene cluster participating in menaquinone biosynthesis [30]. Ashby & Edwards [31] have isolated from a plasmid containing a wild-copy genomic DNA fragment a gene (Coq1) that is able to complement the yeast mutant in coenzyme Q biosynthesis and restore its HexPP synthase activity. This enzyme which has conserved regions characteristic of prenyltransferases, seems to correspond to the larger protein component of the two heterodimeric components of bacterial HexPP synthase. However, it is not known whether the yeast protein acts as HexPP synthase by itself or in association with another gene product similar to the smaller protein component.
Koike-Takeshita, A., Koyama, T. & Ogura, K. (submitted for publication).
474 Heptaprenyl diphosphate synthases
A. Szkopiska
2000
Heptaprenyl diphosphate (HepPP) synthase [EC 2.5.1.30] has been found in Bacillus subtilis which produces exclusively menaquinone with a side chain of 7 isoprene units [32]. It catalyzes the synthesis of (all-E) HepPP by adding four molecules of IPP to FPP but like HexPP synthase it does not synthesize C5 C10 C15 compounds. On the basis of the highly conserved aminoacid sequences of prenyltransferases two genes encoding two protein components of HepPP synthase from Bacillus stearothermophilus were identified [33]. One of the proteins constituting an enzyme system for the synthesis of HepPP has 323 amino-acid residues and shows 32% sequence similarity to the FPP synthase from the same bacterium [34]. This protein designated as component II has highly conserved regions typical of prenyltransferases. The other protein (component I) is composed of 220 amino acids and has no such similarity nor has any similar protein been found within the protein entries in data bases. Therefore, it seems likely that component II carries the active sites for substrate binding and catalysis, while component I plays an auxiliary role. A protein database search for amino-acid sequences similar to that of component II of the HepPP synthase from B. stearothermophilus [33] yielded the GerC3 protein of B. subtilis which is encoded in a cluster of three ORFs, gerC1, gerC2 and gerC3. Two of the gerC products, GerC1 and GerC3, correspond to the two dissociable components I and II which constitute the HepPP synthase from B. stearothermophilus [35].
Octaprenyl diphosphate synthases
homodimeric protein that is functionally active by itself. Analysis of genes in the E. coli chromosome performed by Choi et al. [37] revealed an open-reading frame that showed significant similarity to the ispA gene which encodes FPP synthase of E. coli [38]. Jeong et al. [39] determined the entire sequence of this open reading frame and found a high similarity of the gene product to the HexPP synthase of S. cerevisiae [31] and the GGPP synthases of various organisms. Asai et al. [40] identified the ispB (cel) gene encoding the octaprenyl diphosphate synthase in E. coli. The deduced amino-acid sequence of this enzyme also shows the presence of the seven conserved regions typical of prenyltransferases [34]. Okada et al. [41] proved that E. coli octaprenyl diphosphate synthase (IspB) having a mitochondrial import signal is expressed in S. cerevisiae. Yeast cells produced ubiquinone 8 in addition to the originally existing ubiquinone 6. When IspB was expressed in a S. cerevisiae COQ1 defective strain, IspB complemented the defect of growth on a non-fermentable carbon source.
Nonaprenyl and decaprenyl diphosphate synthases
Bacterial (all-E) octaprenyl diphosphate synthase (forming an 8 isoprene residue polyprenyl chain) has been found and partially purified from E. coli [36]. This enzyme is a
2
(all-E) Nonaprenyl (solanesyl) diphosphate synthase (SPP) [EC 2.5.1.11] was isolated from M. luteus [42] as an enzyme catalyzing chain elongation from GPP up to C45PP. The enzyme is a homodimeric protein [43]. However, it requires a protein factor to maintain efficient catalytic turnover. This factor is a high molecular mass component of a soluble fraction of the same bacterium. Possibly it acts by removing from the active site the hydrophobic products which otherwise would inhibit the reaction. Decaprenyl diphosphate synthase has been found and partially purified from Paracoccus denitrificans [44]. It is also a homodimeric protein utilizing GPP as substrate. Suzuki et al. [45] and Takahashi et al.2 have cloned the
Takahashi, S., Koyama, T., Nishino, T., (submitted for publication).
Vol. 47
Ubiquinone biosynthesis LOCALIZATION OF QUINONE SYNTHESIS
475
gene encoding the decaprenyl diphosphate synthase of the fission yeast Schizosaccharomyces pombe and P. denitrificans, respectively. Both nona- and decaprenyl diphosphate synthases are present in plants. They have been characterized in spinach leaves [47]. As does the bacterial enzyme, they utilize GPP as substrate. In most rat tissues, ubiquinone contains 9 isoprene residues but in brain, spleen and intestine, one-third of the ubiquinone has a decaprenyl side-chain [4850].
Knowledge about the intracellular localization of the ubiquinone synthesizing enzymes is scant. They are believed to be membranebound or membrane associated [56]. It is thought that ubiquinone synthesis in cells of higher eukaryotes reflects the yeast pathway. In plant cells, the precise localization of this biosynthetic pathway is a matter of controversy. Lutke-Brinkhaus et al. [55] and Lutke-
Figure 6. The dynamic formation and dissociation of the ternary complex of the heteromeric components of hexaprenyl diphosphate synthase. The enzyme catalyzes the synthesis of (all-E) hexaprenyl diphosphate by adding three molecules of isopentenyl diphosphate (IPP) to farnesyl diphosphate (FPP) [27].
In all human tissues the predominant side chain of ubiquinones is decaprenol and only small proportion (27%) of nonaprenol is present.
TRANSFER OF POLYPRENYL DIPHOSPHATES TO paraHYDROXYBENZOATE
The E. coli gene UBIA encoding prenyltransferase catalysing the transfer of the isoprenoid side chain to para-hydroxybenzoate has been isolated [51]. The UBIA of E. coli and COQ2 of S. cerevisiae genes have been analyzed and cloned [5254]. Lutke-Brinkhaus et al. [55] have described PHB-polyprenyltransferase activity in plant mitochondria. The enzyme has a very broad substrate specificity.
Brinkhaus and Kleinig [57] found that plant mitochondria purified from potato tubers catalyze several reactions in the biosynthesis of ubiquinones. In contrast, wieewska et al. [47] found nonaprenyl-4-hydroxybenzoate and nonaprenyl-2-methylquinol transferase activities in microsomal and Golgi preparations, and proposed therefore, that ubiquinone biosynthesis occurs in the endoplasmic reticulum-Golgi system, followed by a selective transfer of ubiquinone to the mitochondria. In animal systems the enzymes of the mevalonic acid pathway which supply polyprenyl side chain have a multilocal intracellular distribution. In addition to mitochondria [49] they are present in the endoplasmic reticulum [58], peroxisomes [59], Golgi system, lysosomes, plasma mem-
476
A. Szkopiska
2000
brane [60] and cytosol [61]. According to Dallner and his group [62] ubiquinone synthesis begins in the endoplasmic reticulum and is completed in the Golgi system, after which ubiquinone becomes distributed to other cellular organelles.
REGULATION OF UBIQUINONE SYNTHESIS
The factors influencing the regulation of ubiquinone cellular level are numerous. In view of the complexity of their function, structure and localization of synthesis, ubiquinones might be the subject of another review article. Only a few general points will be mentioned here. Ubiquinone levels are regulated through the mevalonate pathway. Moreover, there is an intricate interplay among the three major biosynthetic products of mevalonate metabolism, i.e. ubiquinone, dolichol and sterol (Fig. 3) [63, 64]. In E. coli the composition of the ubiquinone pool is highly influenced by the degree of oxygen availability: aerobically grown E. coli cells contain significantly more ubiquinone 8. The mechanism of this regulation is not yet shown [65]. In yeast under glucose derepression the level of ubiquinone 6 synthesized increases.
Much less is known about the physiological regulation of ubiquinone level. No data is available concerning plants. In animal systems it has been observed that the ubiquinone level increases upon various forms of oxidative stress (physical exercise, cold adaptation, thyroid hormone treatment) and decreases with age. Relatively little is known about the mechanism involved in biodegradation of ubiquinone. The turnover rate of ubiquinone in various tissues is rather similar, ranging from 50 to 125 h [66]. This is in contrast to cholesterol and dolichol for which the turnover rates are several orders of magnitude higher in the liver than in the brain [67]. The decrease in ubiquinone content with increasing age is consistent with the free radical theory of aging [68], as reflected by an inverse correlation between longevity and peroxide-producing potential in mammalian tissues [69]. This decrease may also account for the age-related increase in the extent of oxidative damage to proteins [70] and DNA [71], the latter especially to mitochondrial DNA [72, 73]. Increase of ubiquinone concentration was found in neurodegenerative conditions in the brain, such as in Alzheimers disease [74] and prion disease in mice [75]. It is not yet clear whether these diseases are related to an alteration in the bioenergetic capacity and/or in the antioxidant status of the tissues concerned.
REFERENCES 1. Crane, F.L., Hatefi,Y., Lester, R.L. & Widmer, C. (1957) Isolation of a quinone from beef heart mitochondria. Biochim. Biophys. Acta 25, 220-221. MEDLINE 2. Wolf, D.E., Hoffman, C.H., Trenner, N.R., Arison, B.H., Shunk, C.H., Linn, B.O., McPherson, J.F. & Folkers, K. (1958) Coenzyme Q. Structure studies on the coenzyme Q group. J. Am. Chem. Soc. 80, 4752-4758. 3. Festenstein, G.N., Heaton, F.W., Lowe, J.S. & Morton, R.A. (1955) A constituent of the unsaponificable portion of the animal tissue lipids (lambdamax 272 m). Biochem. J. 59, 558-566. MEDLINE 4. Genova, M.L., Bonacorsi, E., D'Aurelio, M., Formiggini, G., Narudo, B., Cuccomarino, S., Turi, P., Pich, M., Lenaz, G. & Bovina, C. (1999) Protective effect of exogenous coenzyme Q in rats subjected to partial hepatic ischemia and reperfusion. Biofactors 9, 345-349 MEDLINE 5. Miksovska, J., Schiffer, M., Hanson, D. & Sebban, K. (1999) Proton uptake by bacterial reaction centers: The protein complex responds in a similar manner to the reduction of either quinone acceptor. Proc. Natl. Acad. Sci. U.S.A. 96, 14348-14353. MEDLINE 6. Gillman, I., Clark, T., N. & Manderville, R.A. (1999) Oxidation of ochratoxin A by an Feporphyrin system: Model for enzymatic activation of DNA cleavage. Chem. Res. Toxicol. 12, 1066-1076. MEDLINE 7. Hundal, T., Forsmark, P., Ernster, L. & Andersson, B. (1992) Antioxidant activity of reduced plastoquinone in thylacoid membranes during strong illumination. EBEC Short Reports 7, 6-8. 8. Aiyar, A. & Olson, R. (1964) Biosynthesis of the benzoquinone ring of coenzyme Q9 in the rat. Federation Proc. 23, 425. 9. Olson, R. (1965) Anabolism of the coenzyme Q family and their biological activities. Federation Proc. 24, 85-92. 10. Parson, W. & Rudney, H. (1965) The biosynthesis of ubiquinone and rhodoquinone from p-hydroxybenzoate and p-hydroxybenzaldehyde in Rhodospirillum rubrum. J. Biol. Chem. 240, 1855-1863. MEDLINE 11. Olson, R., Dialameh, G. & Bentley, R. (1961) Ciba Fundation Symposium on Quinones in Electron Transport (Welstenholme, G. & O'Connor, C., eds.) London. 12. Ramasarma, T. (1985) in Coenzyme Q Biochemistry, Bioenergetics and Clinical Applications of Ubiquinone (Lenaz, G., ed.) pp. 67-81, Wiley, New York. 13. Kang, D., Takeshigate, K., Isobe, R. & Minakami, S. (1991) Evidence that the decarboxylation reaction occurs before the first methylation in ubiquinone biosynthesis in rat liver mitochondria. Eur. J. Biochem. 198, 599-605. MEDLINE 14. Wu, G., Wiliams, H., Zamanian, M., Gibson, F. & Poole, R. (1992) Isolation and characterization of Escherichia coli mutants affected in aerobic respiration: the cloning and nucleotide sequence of ubiG; identification of an S-adenosylmethionine binding motif in protein, RNA and small-molecule methyltransferases. J. Gen. Microbiol. 138, 2101- 2111. MEDLINE 15. Clarke, C., Wiliams, W. & Teruya, J. (1991) Ubiquinone biosynthesis in Saccharomyces cerevisiae. Isolation and sequence of coq3, the 3,4-dihydroxy-5-hexaprenylbenzoate methyltransferase gene. J. Biol. Chem. 266, 16366-16644. MEDLINE 16. Nakahigashi, K., Miyamoto, K., Nishimura, K. & Inokuchi, H. (1992) Isolation and characterization of a light sensitive mutant of Escherichia coli K-12 with a mutation in a gene that is required for the biosynthesis of ubiquinone. J. Bacteriol. 174, 7352-7359. MEDLINE 17. Huang, J., He, Z. & Wiegel, J. (1999) Cloning, characterization, and expression of a novel gene encoding a reversible 4-hydroxybenzoate decarboxylase from Clostridium hydroxybenzoicum. J. Bacteriol. 181, 5119-5122. MEDLINE 18. Marbois, B., Hsu, A., Pillai, R., Colicelli, J. & Clarke, C. (1994) Cloning of a rat cDNA encoding dihydroxypolyprenylbenzoate methyltransferase by functional complementation of a Saccharomyces cerevisiae mutant deficient in ubiquinone biosynthesis. Gene 138, 213-217. MEDLINE 19. Avelange-Macherel, M.-H. & Joyard, J. (1998) Cloning and functional expression of AtCOQ3, the Arabidopsis homologue of the yeast COQ3 gene, encoding a methyltransferase from plant mitochondria involved in ubiquinone biosynthesis. Plant J. 14, 203-213. MEDLINE 20. Lynen, F., Eggerer, H., Henning, U. & Kessel, I. (1958) Angew. Chem. 70, 738-742. 21. Chaykin, S., Law, J., Philips, A., Tchen, T. & Bloch, T. (1958) Phosphorylated intermediates in the synthesis of squalene. Proc. Natl. Acad. Sci. U.S.A. 44, 998-1004. MEDLINE 22. Rohmer, M., Knani, M., Simonin, P., Sutter, B. & Sahm, H. (1993) Isoprenoid biosynthesis in bacteria: A novel pathway for the early steps leading to isopentenyl diphosphate. Biochem. J. 295, 517-524. MEDLINE 23. Rohmer, M., Seeman, M., Horbach, S., Bringer-Mayer, S. & Sahm, H. (1996) Isopentenol pyrophosphate isomerase. J. Am. Chem. Soc. 118, 2564-2566. 24. Agranoff, B., Eggerer, H., Henning, U. & Lynen, F. (1959) Isopentenol pyrophosphate isomerase. J. Am. Chem. Soc. 81, 1254-1255. 25. Tarshis, L., Proteau, P., Kellogg, B., Sacchettini, J. & Poulter, C. (1996) Regulation of product chain length by isoprenyl diphosphate synthases. Proc. Natl. Acad. Sci. U.S.A. 93, 15018-15023. MEDLINE 26. Jeffries, L., Cawthorn, M., Harris, M., Diplock, A., Green, J. & Price, S. (1967) Distribution of menaquinones in aerobic Micrococcaceae. Nature 215, 257-259. MEDLINE 27. Fujii, H., Koyama, T. & Ogura, K. (1982) Hexaprenyl pyrophosphate synthetase from Micrococcus luteus B-P 26. J. Biol. Chem. 257, 14610-14612. MEDLINE 28. Shimizu, N., Koyama, T. & Ogura, K. (1998) Molecular cloning, expression, and characterization of the two genes encoding the two essential protein components of Micrococcus luteus B-P 26 hexaprenyl diphosphate synthase. J. Bacteriol. 180, 1578-1581. MEDLINE 29. Reference in the text as footnote 1. 30. Koike-Takeshita, A., Koyama, T. & Ogura, K. (1997) Identification of a novel gene cluster participating in menaquinone (Vitamin K2) biosynthesis. J. Biol. Chem. 272, 12380- 12383. MEDLINE 31. Ashby, M. & Edwards, P. (1990) Elucidation of deficiency in two yeast coenzyme Q mutants. J. Biol. Chem. 265, 13157-13164. MEDLINE 32. Takahashi, I., Ogura, K. & Seto, S. (1980) Heptaprenyl pyrophosphate synthetase from Bacillus subtilis J. Biol. Chem. 255, 4539- 4543. MEDLINE 33. Koike-Takeshita, A., Koyama, T., Obata, S. & Ogura, K. (1995) Molecular cloning and nucleotide sequence of the genes for two essential proteins constituting a novel enzyme for heptaprenyl diphosphate synthesis. J. Biol. Chem. 270, 18396-18400. MEDLINE 34. Koyama, T., Obata, S., Osabe, M., Takeshita, A., Yokoyama, K., Uchida, M., Nishino, T. & Ogura, K. (1993) Thermostable farnesyl diphosphate synthase of Bacillus stearothermophilus: Molecular cloning, sequence determination, overproduction, and purification. J. Biochem. 113, 355-363. MEDLINE 35. Zang, Y.-W., Koyama, T. & Ogura, K. (1997) Two cistrons of the gerC operon of Bacillus subtilis encode the two subunits of heptaprenyl diphosphate synthase. J. Bacteriol. 179, 1417-1419. MEDLINE 36. Fujisaki, S., Nishino, T. & Katsuki, H. (1986) Isoprenoid synthesis in Escherichia coli. Separation and partial purification of four enzymes involved in the synthesis. J. Biochem. 99, 1327-1337. MEDLINE 37. Choi, Y., Nishida, T., Kawamukai, M., Utsumi, R., Sakai, H. & Kamono, T. (1989) Cloning and sequencing of an Escherichia coli gene nlp, highly homologous to the ner genes of bacteriophages Mu and D108. J. Bacteriol. 171, 5222-5225. MEDLINE 38. Fujisaki, S., Hara, H., Nishimura, Y., Horiuchi, K. & Nishino, T. (1990) Cloning and nucleotide sequence of the ispA gene responsible for farnesyl diphosphate synthase activity in Escherichia coli. J. Biochem. 108, 995-1000. MEDLINE 39. Jeong, J., Kitakawa, M. & Isono, K. (1993) Cloning and nucleotide sequencing of the genes, rpIU and rpmA, for ribosomal proteins L21 and L27 of Escherichia coli. DNA Sequence 4, 59-67. 40. Asai, K., Fujisaki, S., Nishimura, Y., Nishino, T., Okada, K., Nakagawa, T., Kawamukai, M. & Matsuda, H. (1994) The identification of Escherichia coli ispB (cel) gene encoding the octaprenyl diphosphate synthase. Biochem. Biophys. Res. Commun. 202, 340-345. MEDLINE 41. Okada, K., Suzuki, K., Kamiya, Y., Zhu, X., Fujisaki, S., Nishimura, Y., Nishino, T., Nakagawa, T., Kawamukai, M. & Matsuda, H. (1996) Polyprenyl diphosphate synthase essentially defines the length of the side chain of ubiquinone. Biochim. Biophys. Acta 1302, 217-223. MEDLINE 42. Sagami, H., Ogura, K. & Seto, S. (1977) Solanesyl pyrophosphate synthase from Micrococcus lysodeikticus. Biochemistry 16, 4616-4622. MEDLINE 43. Ohnuma, S., Koyama, T. & Ogura, K. (1991) Purification of solanesyl-diphosphate synthase from Micrococcus luteus. J. Biol. Chem. 266, 23706-23713. MEDLINE 44. Ishi, K., Sagami, H. & Ogura, K. (1985) Decaprenyl pyrophosphate synthetase from Paracoccus denitrificans. Biochim. Biophys. Acta 835, 291-297. MEDLINE 45. Suzuki, K., Okada, K., Kamiya, Y., Zhu, X., Nakagawa, T., Kawamukai, M. & Matsuda, H. (1997) Analysis of the decaprenyl diphosphate synthase (dps) gene in fission yeast suggests a role of ubiquinone as an antioxidant. J. Biochem. 121, 496-505. MEDLINE 46. Reference in the text as footnote 2. 47. Swiezewska, E., Dallner, G., Andersson, B. & Ernster, L. (1993) Biosynthesis of ubiquinone and plastoquinone in the endoplasmic reticulum-Golgi membranes of spinach leaves. J. Biol. Chem. 268, 1494-1499. MEDLINE 48. Teclebrahn, H., Olsson, J., Swiezewska, E. & Dallner, G. (1993) Biosynthesis of the side chain of ubiquinone: Trans-prenyltransferase in rat liver microsomes. J. Biol. Chem. 268, 23081-23086. MEDLINE 49. Runquist, M., Ericsson, J., Thelin, A., Chojnacki, T. & Dallner, G. (1994) Isoprenoid biosynthesis in rat liver mitochondria. Studies on farnesyl pyrophosphate synthase and trans-prenyltransferase. J. Biol. Chem. 269, 5804-5809. MEDLINE 50. Drabikowska, A. (1969) Biosynthesis of ubiquinone in animal tissues. Post. Biochem. 15, 397-410 (in Polish). 51. Wu, G., Wiliams, H., Gibson, F. & Poole, K. (1993) Mutants of Escherichia coli affected in respiration: The cloning and nucleotide sequence of ubiA, encoding the membrane- bound p-hydroxybenzoate:octaprenyltransferase. J. Gen. Microbiol. 139, 1795-1805. MEDLINE 52. Ashby, M., Kutsunai, S., Ackerman, S., Tzagoloff, A. & Edwards, P. (1992) COQ2 is a candidate for the structural gene encoding para-hydroxybenzoate:polyprenyltransferase. J. Biol. Chem. 267, 4128-4136. MEDLINE 53. Siebert, M., Bechthold, A., Melzer, M., May, U., Berger, U., Schroder, G., Schroder, J., Severin, K. & Heide, L. (1992) Cloning of the genes coding for chorismate pyruvate-lyase and 4-hydroxybenzoate octaprenyl transferase from Escherichia coli. FEBS Lett. 307, 347-350. MEDLINE 54. Melzer, M. & Heide, L. (1994) Characterization of polyprenyldiphosphate:4-hydroxybenzoate polyprenyltransferase from Escherichia coli. Biochim. Biophys. Acta 1212, 93-102. MEDLINE 55. Lutke-Brinkhaus, F., Liedvogel, B. & Kleinig, H. (1984) On the biosynthesis of ubiquinones in plant mitochondria. Eur. J. Biochem. 141, 537-541. MEDLINE 56. Leppik, R., Stroobant, P., Shineberg, B., Young, I. & Gibson, F. (1976) Membrane-associated reactions in ubiquinone biosynthesis. Biochim. Biophys. Acta 428, 146-156. MEDLINE 57. Lutke-Brinkhaus, F. & Kleinig, H. (1987) Ubiquinone biosynthesis in plant mitochondria. Methods Enzymol. 148, 486-490. MEDLINE 58. Ericsson, J., Runquist, M., Thelin, A., Anderson, M., Chojnacki, T. & Dallner (1993) Distribution of prenyltransferases in rat tissues. J. Biol. Chem. 268, 832-838. MEDLINE 59. Aboushadi, N. & Krisans, S. (1998) Analysis of isoprenoid biosynthesis in peroxisomal-deficient Pex2 CHO cell lines. J. Lipid Res. 39, 1781-1791. MEDLINE 60. Kalen, A., Norling, B., Appelkvist, E.-L. & Dallner, G. (1987) Ubiquinone biosynthesis by the microsomal fraction from rat liver. Biochim. Biophys. Acta 926, 70-78. MEDLINE 61. Engfelt, W., Shackelford, E., Aboushadi, N., Jessani, N., Masuda, K., Paton, V., Keller, G. & Krisans, K. (1997) The characterization of UT-2 cells. The induction of peroxisomal 3-hydroxy-3-methylglutaryl coenzyme A. J. Biol. Chem. 272, 24579-24587. MEDLINE 62. Elmberger, G., Kalen, A., Brunk, U. & Dallner, G. (1989) Discharge of newly synthesized dolichol and ubiquinone with lipoproteins to rat liver perfusate and to the bile. Lipids 24, 919-930. MEDLINE 63. Szkopinska, A., Swiezewska, E. & Karst, F. (2000) The regulation of activity of main mevalonic acid pathway enzymes: Farnesyl diphosphate synthase, 3-hydroxy-3-methylglutaryl-CoA reductase and squalene synthase in yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 267, 473-477. MEDLINE 64. Grabowska, D., Karst, F. & Szkopinska, A. (1998) Effect of squalene synthase gene disruption on synthesis of polyprenols in Saccharomyces cerevisiae. FEBS Lett. 434, 406-408. MEDLINE 65. Meganathan, A. (1996) Biosynthesis of the isoprenoid quinones, menaquinone (vitamin K2 ) and ubiquinone (coenzymeQ); in Escherichia coli and Salmonella: Cell. Mol. Biol. (Neidhardt F.C., et al.) pp. 642-656, American Society for Microbiology, Washington. 66. Thelin, A., Schedin, S. & Dallner, G. (1992) Half-life of ubiquinone-9 in rat tissues. FEBS Lett. 313, 118-120. MEDLINE 67. Andersson, M., Elmberger, P., Edlund, C. Kristensson, K. & Dallner, G. (1990) Rates of cholesterol, ubiquinone, dolichol, and dolichyl-P biosynthesis in rat brain slices. FEBS Lett. 269, 15-18. MEDLINE 68. Harman, D. (1988) Free radicals in aging. Mol. Cell. Biochem. 84, 55-61. MEDLINE 69. Cutler, R. (1985) Peroxide-producing potential of tissues: Inverse correlation with longevity of mammalian species. Proc. Natl. Acad. Sci. U.S.A. 82, 4798-4802. MEDLINE 70. Stadtman, E. & Oliver, C. (1991) Metal-catalyzed oxidation of proteins. Physiological consequences. J. Biol. Chem. 266, 2005-2008. MEDLINE 71. Linnane, A., Marzuki, S., Ozawa, T. & Tanaka, M. (1989) Mitochondrial DNA mutations as an important contributor to ageing and degenerative diseases. Lancet 25, 642-645. MEDLINE 72. Ozawa, T. (1997) Genetic and functional changes in mitochondria associated with aging. Physiol. Rev. 77, 425-464. MEDLINE 73. Lenaz, G., Cavazzoni, M., Genova, M., D'Aurelio, M., Merlo, M., Palloti, F., Formiggini, G., Marchetti, M., Parenti, G. & Bovina, C. (1998) Oxidative stress, antioxidant defences in aging. BioFactors 8, 195-204. MEDLINE 74. Soderberg, M., Edlund, K., Kristensson, K. & Dallner, G. (1990) Lipid compositions of different regions of the human brain during aging. J. Neurochem. 54, 415-423. MEDLINE 75. Ericsson, J. & Dallner, G. (1993) in Subcellular Biochemistry (Borgese, N. & Harris, J., eds.) vol. 21, pp. 229-272, Plenum, New York. 76. Grunler, J., Ericsson, J. & Dallner, G. (1994) Branch-point reactions in the biosynthesis of cholesterol, dolichol, ubiquinone and prenylated proteins. Biochim. Biophys. Acta 1212, 259-277. MEDLINE
Você também pode gostar
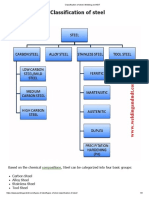
- Classification of Steel - Welding and NDTDocumento3 páginasClassification of Steel - Welding and NDTAshif Iqubal100% (1)
- Ethem Murat Arsava (Eds.) - Nutrition in Neurologic Disorders - A Practical Guide (2017, Springer International Publishing)Documento209 páginasEthem Murat Arsava (Eds.) - Nutrition in Neurologic Disorders - A Practical Guide (2017, Springer International Publishing)riddhiAinda não há avaliações
- Wa0044 PDFDocumento337 páginasWa0044 PDFYamilAinda não há avaliações
- Palm Based Non Hydrogenated Creamer PDFDocumento4 páginasPalm Based Non Hydrogenated Creamer PDFbellesuperAinda não há avaliações
- ENZYME KINETICSDocumento11 páginasENZYME KINETICSDianne Villanueva100% (1)
- Column and Thin Layer ChromatographyDocumento5 páginasColumn and Thin Layer Chromatographymarilujane80% (5)
- Ubiquinone Biosynthesis of QuinoneDocumento12 páginasUbiquinone Biosynthesis of QuinoneFrancisco EspinosaAinda não há avaliações
- Gene Encoding Isopentenyl Diphosphate Isomerase: Escherichia Coli Open Reading Frame 696 Is Idi, A NonessentialDocumento6 páginasGene Encoding Isopentenyl Diphosphate Isomerase: Escherichia Coli Open Reading Frame 696 Is Idi, A NonessentialHuong TranAinda não há avaliações
- CoQ10-Biosynthesis-1-s2.0-S0005272816300573-mainDocumento6 páginasCoQ10-Biosynthesis-1-s2.0-S0005272816300573-maingjlynxAinda não há avaliações
- Anthocyanin Synthesis in Maize Aleurone Tissue: Conek@missouri - EduDocumento2 páginasAnthocyanin Synthesis in Maize Aleurone Tissue: Conek@missouri - EduMonica Cabezas DipazAinda não há avaliações
- Research Project: Prof. Dr. Rehab HamedDocumento13 páginasResearch Project: Prof. Dr. Rehab HamedHassaninElaradyAinda não há avaliações
- Biotin Synthesis in Higher Plants: Purification and Characterization ofDocumento5 páginasBiotin Synthesis in Higher Plants: Purification and Characterization ofmyfreepaysite321Ainda não há avaliações
- Insights Into Phycoerythrobilin Biosynthesis Point Toward Metabolic ChannelingDocumento9 páginasInsights Into Phycoerythrobilin Biosynthesis Point Toward Metabolic ChannelingAvani KaushalAinda não há avaliações
- Chloroplast Import of Four Carotenoid Biosynthetic EnzymesDocumento9 páginasChloroplast Import of Four Carotenoid Biosynthetic EnzymesNithin RAinda não há avaliações
- Seminario 1-2022-1Documento13 páginasSeminario 1-2022-1Elias Gerardo Pardo LlamoccaAinda não há avaliações
- Silva Et Al. 2021Documento11 páginasSilva Et Al. 2021Davi OliveiraAinda não há avaliações
- Structural and Biological Aspects of Carotenoid CleavageDocumento13 páginasStructural and Biological Aspects of Carotenoid CleavageMinh Chau NguyenAinda não há avaliações
- Alkaloids and OthersDocumento24 páginasAlkaloids and OtherszakybaaAinda não há avaliações
- Bacterial Phospholipase A: Structure and Function of An Integral Membrane PhospholipaseDocumento11 páginasBacterial Phospholipase A: Structure and Function of An Integral Membrane PhospholipaseDiana SahoneroAinda não há avaliações
- Botryococcus Braunii, Race B: Isolation and Characterization of Two Squalene Epoxidase Genes FromDocumento14 páginasBotryococcus Braunii, Race B: Isolation and Characterization of Two Squalene Epoxidase Genes FromCarlos A. PonceeAinda não há avaliações
- Redox InfoDocumento11 páginasRedox InfoCasandra Valencia OjedaAinda não há avaliações
- Mitochondrial Diseases and Genetic Defects of ATP SynthaseDocumento6 páginasMitochondrial Diseases and Genetic Defects of ATP SynthasewlymjzdwAinda não há avaliações
- (Department of Zoology,: P-ApocarotenalsDocumento1 página(Department of Zoology,: P-ApocarotenalsKrrliveAinda não há avaliações
- Phosphorylation of Enoyl-Acyl Carrier Protein Reductase Inha Impacts Mycobacterial Growth and SurvivalDocumento12 páginasPhosphorylation of Enoyl-Acyl Carrier Protein Reductase Inha Impacts Mycobacterial Growth and SurvivalNini FarmAinda não há avaliações
- Metabolic Engineering of Cyanobacteria For Ethanol ProductionDocumento8 páginasMetabolic Engineering of Cyanobacteria For Ethanol ProductionPatrick FuAinda não há avaliações
- міоінозитолDocumento10 páginasміоінозитолDaryna DarynaAinda não há avaliações
- Redox BiologyDocumento13 páginasRedox BiologyLin LIAinda não há avaliações
- ATP Synthesis and Storage: Original ArticleDocumento15 páginasATP Synthesis and Storage: Original ArticleRiley RilanAinda não há avaliações
- Association Between Genetic Variants in The Coenzyme Q Metabolism and Coenzyme Q Status in HumansDocumento7 páginasAssociation Between Genetic Variants in The Coenzyme Q Metabolism and Coenzyme Q Status in HumansLUCIENE CRISTALDO ALBUQUERQUEAinda não há avaliações
- Hong 2005Documento11 páginasHong 2005tea.zakicAinda não há avaliações
- Enzymatic Total Synthesis of Gibberellin A4Documento8 páginasEnzymatic Total Synthesis of Gibberellin A4ranukahewageAinda não há avaliações
- Vitamin K Biosynthesis: A. MenaquinoneDocumento4 páginasVitamin K Biosynthesis: A. MenaquinoneArslan Muhammad EjazAinda não há avaliações
- Chemistry of Protein Kinases and Dephosphorylases: Advanced ArticleDocumento8 páginasChemistry of Protein Kinases and Dephosphorylases: Advanced ArticleazzaassAinda não há avaliações
- Ajikumar 2010Documento6 páginasAjikumar 2010Jonathan DiasAinda não há avaliações
- Topic 3.2Documento33 páginasTopic 3.2Salna Susan AbrahamAinda não há avaliações
- Þ CC) C' (CC& +C++C: CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCDocumento12 páginasÞ CC) C' (CC& +C++C: CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCgauravAinda não há avaliações
- Ousalem Et Al 2023Documento46 páginasOusalem Et Al 2023pampetraAinda não há avaliações
- High-Level Production of Recombinant Arenicola Marina Globin Chains in Escherichia Coli: A New Generation of Blood SubstituteDocumento12 páginasHigh-Level Production of Recombinant Arenicola Marina Globin Chains in Escherichia Coli: A New Generation of Blood SubstituteIstván PortörőAinda não há avaliações
- Self-Assembled Protein-Enzyme Nanoflower-Based Fluorescent Sensing For Protein BiomarkerDocumento8 páginasSelf-Assembled Protein-Enzyme Nanoflower-Based Fluorescent Sensing For Protein BiomarkerAlvaro Matus CAinda não há avaliações
- Manipulation of Malic Enzyme in Saccharomyces Cerevisiae For Increasing NADPH ProductionDocumento12 páginasManipulation of Malic Enzyme in Saccharomyces Cerevisiae For Increasing NADPH ProductionJesús Miguel Jacinto NavaAinda não há avaliações
- COQ7 phenotipic, molecular functioningDocumento13 páginasCOQ7 phenotipic, molecular functioningLaura A M MAinda não há avaliações
- FotosintesisDocumento6 páginasFotosintesisNorma YantiAinda não há avaliações
- Fotosintesis PDFDocumento6 páginasFotosintesis PDFNorma YantiAinda não há avaliações
- Artículo RehabilitaciónDocumento10 páginasArtículo RehabilitaciónWilliam rodriguez ramosAinda não há avaliações
- Bacterial Phenylalanine and Phenylacetate Catabolic Pathway RevealedDocumento6 páginasBacterial Phenylalanine and Phenylacetate Catabolic Pathway RevealedKarla KetiAinda não há avaliações
- Inorganic Polyphosphates in Mitochondria Review 2010Documento7 páginasInorganic Polyphosphates in Mitochondria Review 2010Baranov MaximAinda não há avaliações
- Figuras Insight Into The Evolution of Microbial Metabolism From The Deep-Branching Bacterium, Thermovibrio AmmonificansDocumento14 páginasFiguras Insight Into The Evolution of Microbial Metabolism From The Deep-Branching Bacterium, Thermovibrio AmmonificansIara GAinda não há avaliações
- Biochemical and Biophysical Research CommunicationsDocumento5 páginasBiochemical and Biophysical Research CommunicationsFernando P. Molina-HerediaAinda não há avaliações
- Caracterización de La Olivetol Sintasa, Una Policétido Sintasa Implicada Supuestamente en La Ruta Biosintética de Los CannabinoidesDocumento6 páginasCaracterización de La Olivetol Sintasa, Una Policétido Sintasa Implicada Supuestamente en La Ruta Biosintética de Los CannabinoidesAnonymous 9oH7eQ2TAinda não há avaliações
- First Report: Yersinia Enterocolitica Recovered From Canine TonsilsDocumento5 páginasFirst Report: Yersinia Enterocolitica Recovered From Canine TonsilsMarcela TapiasAinda não há avaliações
- Babesia Canis Rossi: Characterization and Molecular Cloning of An Adenosine Kinase FromDocumento7 páginasBabesia Canis Rossi: Characterization and Molecular Cloning of An Adenosine Kinase FromSoare MarianAinda não há avaliações
- TMP 2 FEDocumento2 páginasTMP 2 FEFrontiersAinda não há avaliações
- Peroxisomal β-oxidation-A metabolic pathway with multiple functionsDocumento14 páginasPeroxisomal β-oxidation-A metabolic pathway with multiple functionsRoy WilsonAinda não há avaliações
- 5 Complementation of Apolipoprotein B mRNA Editing by Human Liver Accompanied by Secretion of Apolipoprotein B48 J. Biol. Chem.-1994-Giannoni-5932-6Documento5 páginas5 Complementation of Apolipoprotein B mRNA Editing by Human Liver Accompanied by Secretion of Apolipoprotein B48 J. Biol. Chem.-1994-Giannoni-5932-6Federico GiannoniAinda não há avaliações
- Cloning and expressing cellulase gene in silkworm using baculovirus systemDocumento8 páginasCloning and expressing cellulase gene in silkworm using baculovirus systemBhaskar RoyAinda não há avaliações
- Unesco - Eolss Sample Chapters: Bioplastic and Biopolymer ProductionDocumento10 páginasUnesco - Eolss Sample Chapters: Bioplastic and Biopolymer ProductionAliAliAinda não há avaliações
- Bioorganic & Medicinal ChemistryDocumento6 páginasBioorganic & Medicinal ChemistryRuthaiwan KongcharoenAinda não há avaliações
- Microbial Production of Biodegradable Polymers and Their Role in Cardiac Stent DevelopmentDocumento11 páginasMicrobial Production of Biodegradable Polymers and Their Role in Cardiac Stent Development2begeniusAinda não há avaliações
- Asembly VLDLDocumento4 páginasAsembly VLDLCornel GheraseAinda não há avaliações
- Art:10 1007/BF00173913Documento8 páginasArt:10 1007/BF00173913Leonardo SakamotoAinda não há avaliações
- 1 s2.0 0014579394009325 MainDocumento5 páginas1 s2.0 0014579394009325 MainMuhammad Andika PerdanaAinda não há avaliações
- 1 s2.0 S0005273614002454 MainDocumento8 páginas1 s2.0 S0005273614002454 MainAlex WasabiAinda não há avaliações
- A Minimal System Allowing Tubulation With Molecular Motors Pulling On Giant LiposomesDocumento6 páginasA Minimal System Allowing Tubulation With Molecular Motors Pulling On Giant LiposomesAbhishek KansalAinda não há avaliações
- Purine Metabolism PathwaysDocumento29 páginasPurine Metabolism Pathwaystrinitysugumar0% (1)
- Recent Advances in Polyphenol ResearchNo EverandRecent Advances in Polyphenol ResearchKumi YoshidaAinda não há avaliações
- Molecules: ISSN 1420-3049Documento25 páginasMolecules: ISSN 1420-3049Jesus LirioAinda não há avaliações
- Ceramic AsDocumento2 páginasCeramic AsJesus LirioAinda não há avaliações
- Solanum Mammosum LDocumento21 páginasSolanum Mammosum LJesus LirioAinda não há avaliações
- Antibacterial Activity of Tannins From The Leaves of Solanum Trilobatum Linn - Feb09dos PDFDocumento3 páginasAntibacterial Activity of Tannins From The Leaves of Solanum Trilobatum Linn - Feb09dos PDFarca_77@Ainda não há avaliações
- Transfer en CIA de Electrones en BacteriasDocumento6 páginasTransfer en CIA de Electrones en BacteriasJesus LirioAinda não há avaliações
- LOVely EnzymesDocumento10 páginasLOVely EnzymesJesus LirioAinda não há avaliações
- Brittany L. Hayes - Recent Advances in Microwave - Assisted SynthesisDocumento11 páginasBrittany L. Hayes - Recent Advances in Microwave - Assisted SynthesisnnnnjwAinda não há avaliações
- Plastics: Name: Taaha Muzaffar Imam ROLL NO.: 19011AA002 Sem/Sec: Ii/A Branch: B.Arch College: Spa' JnafauDocumento5 páginasPlastics: Name: Taaha Muzaffar Imam ROLL NO.: 19011AA002 Sem/Sec: Ii/A Branch: B.Arch College: Spa' JnafauTaaha Muzaffar ImamAinda não há avaliações
- Rambutan SunscreenDocumento7 páginasRambutan SunscreenAndrea Alvarado RoAinda não há avaliações
- Puresilk Salt ChlorinatorDocumento10 páginasPuresilk Salt Chlorinatornike_y2kAinda não há avaliações
- Experiment 2: Brinell Hardness TestDocumento5 páginasExperiment 2: Brinell Hardness TestseifAinda não há avaliações
- 1 ElectrochemistryExercise PDFDocumento46 páginas1 ElectrochemistryExercise PDFDivyanshi TiwaryAinda não há avaliações
- Prepared by Miss Unnati .M. Patel F.Y.M Pharm (Pqa) by DR A.D.Kulkerni Mpharm (Pharmaceutics)Documento29 páginasPrepared by Miss Unnati .M. Patel F.Y.M Pharm (Pqa) by DR A.D.Kulkerni Mpharm (Pharmaceutics)Fatima VessaliusAinda não há avaliações
- OVERVIEW (4 Points) : CH116 General and Organic Principles LabDocumento4 páginasOVERVIEW (4 Points) : CH116 General and Organic Principles Labapi-557329548Ainda não há avaliações
- Company Profile 4 April - CompressedDocumento10 páginasCompany Profile 4 April - CompressedPhilip PhilipsAinda não há avaliações
- Glyphosate Goker MSDS 1Documento7 páginasGlyphosate Goker MSDS 1Bima SitorusAinda não há avaliações
- Moisture RemovalDocumento9 páginasMoisture RemovalRafi AkbarAinda não há avaliações
- Content of The Dossier For Chemical Purity and Microbiological QualityDocumento23 páginasContent of The Dossier For Chemical Purity and Microbiological QualityjdemelloAinda não há avaliações
- Solid Hollow Plug PDFDocumento3 páginasSolid Hollow Plug PDFSandra DevannyAinda não há avaliações
- Chap 8 Ques - AnsDocumento11 páginasChap 8 Ques - AnsHaley WillhelmAinda não há avaliações
- Certificado de Calidad Caps SCH-40Documento1 páginaCertificado de Calidad Caps SCH-40Jesus CondoriAinda não há avaliações
- BooCax Disinfection Solution For Beijing 2022 Winter OlympicsDocumento21 páginasBooCax Disinfection Solution For Beijing 2022 Winter OlympicsRobotics BoocaxAinda não há avaliações
- GATE-Architecture Sample QuestionsDocumento6 páginasGATE-Architecture Sample QuestionsCharan ReddyAinda não há avaliações
- Year 8 Science End of Term 1 Test: AnswerDocumento11 páginasYear 8 Science End of Term 1 Test: Answerchan myaeAinda não há avaliações
- 1 s2.0 S1687157X17300288 MainDocumento11 páginas1 s2.0 S1687157X17300288 Mainnagi jainAinda não há avaliações
- Tandem Dyes For Flow Cytometry, Quality Concerns, Beckman CoDocumento2 páginasTandem Dyes For Flow Cytometry, Quality Concerns, Beckman CocandiddreamsAinda não há avaliações
- Fmi Unit 2Documento86 páginasFmi Unit 2Pranav vigneshAinda não há avaliações
- Human Respiratory System Based On Law of ThermodynamicsDocumento9 páginasHuman Respiratory System Based On Law of ThermodynamicsfatimahAinda não há avaliações
- Mass-Transfer Diffusion Coefficients in Binary Systems: AppendixDocumento3 páginasMass-Transfer Diffusion Coefficients in Binary Systems: AppendixAnisaAinda não há avaliações
- Ekatalog 2023 Sulsel RajawaliDocumento50 páginasEkatalog 2023 Sulsel RajawaliSafria HamzaAinda não há avaliações