Escolar Documentos
Profissional Documentos
Cultura Documentos
Plasmid Identification Experiment Analysis
Enviado por
api-2536039440 notas0% acharam este documento útil (0 voto)
15 visualizações4 páginasTítulo original
plasmid identification experiment analysis
Direitos autorais
© © All Rights Reserved
Formatos disponíveis
DOCX, PDF, TXT ou leia online no Scribd
Compartilhar este documento
Compartilhar ou incorporar documento
Você considera este documento útil?
Este conteúdo é inapropriado?
Denunciar este documentoDireitos autorais:
© All Rights Reserved
Formatos disponíveis
Baixe no formato DOCX, PDF, TXT ou leia online no Scribd
0 notas0% acharam este documento útil (0 voto)
15 visualizações4 páginasPlasmid Identification Experiment Analysis
Enviado por
api-253603944Direitos autorais:
© All Rights Reserved
Formatos disponíveis
Baixe no formato DOCX, PDF, TXT ou leia online no Scribd
Você está na página 1de 4
Plasmid Identification Experiment Analysis
Plasmid Code: 5832D84
By: Michael Shields
In this experiment, I, Michael Shields, identified an unknown plasmid using restriction
enzymes and gel electrophoresis. A plasmid is a DNA molecule that can be manipulated easily.
1
A restriction enzyme is simply an enzyme capable of cutting DNA in a specific location.
2
Gel
electrophoresis is a method of cutting DNA, according to size. Through this process the
molecules to be separated are pushed by an electrical field through a gel that contains small
pores.
3
The DNA then travels at different speeds according to its size, (small pieces go faster
and farther while big pieces go slower, and not as far.) The goal of this experiment was to
analyze and evaluate the type of plasmid that I was given (pAMP, pKAN, or pBLU), as well as to
do so by utilizing my own methods and processes. This goal is particularly important to
understanding biotechnology lab techniques because it was my specific techniques and
research, instead of simply copying someone elses work. My strategy during this experiment
was to utilize a very specific set of restriction enzymes so as to make data analysis easy.
The methods for this project were fairly simple. I first ascertained the restriction
enzymes that Id be using, after that I mixed them with the plasmid assigned to me (code name:
5832D84), according to my recipe. My recipe for this plasmid mixture was 3.3microliters of the
DNA, 1 microliter of the enzyme (either 1microliter of EcoRI, or 1microliter of Ndel and PstI),
and adding 2 microliters of buffer (3.1) and bring the mix to a volume of 20 microliters with
deionized water (dH
2
O). The concentration of the solution was given (150ng/ml). Next, I
incubated the mixture in a heat bath (about 30
o
Celsius) for one hour, while that was happening
I mixed together an agarose gel (0.8% agarose, made by combining 0.4g of agarose with 50mL
of dH
2
O and bringing to volume 150mL). After the plasmid had incubated I ran the DNA in a gel
electrophoresis chamber, with two ladders, for 1.5 hours, at 130 volts. The source of my ladder
was New England Biology Labs, as well as my enzymes and original plasmid. I then removed the
gel, photographed it, and analyzed the data. To analyze the data, I measured the lengths of the
ladder cuts and compared those to the lengths of my own DNAs cuts. I was able to predict this
contrast with the use of an online software
4
.
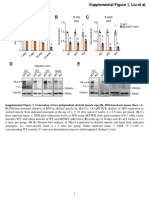
The starting concentration of my plasmid was 150ng/ml. Below, are the two pictures of
my gel, and here is a graph of my Ladder Migration, it shows that I have my cuts (from the
plasmid) on track with how my ladder was supposed to cut.
This is a table showing my results from (
4
) this is the data that was used to find what plasmid
was being analyzed, as well as my plasmid (X) and allowed for the results to be found.
Single Digest Double Digest
Plasmid Enzyme: EcoRI Plasmid Enzyme: Ndel PstI Both (Ndel & PstI)
pAMP 4539 pAMP 4539 4539 3134, 1405
pKAN 4194 pKAN 4194 3271, 923 2635, 923, 636
pBLU 5437 pBLU 5437 3725, 1134, 197 2498, 1316, 926, 197
y = 30.483e
-0.117x
R = 0.9704
0
5
10
15
20
25
30
35
0 2 4 6 8 10 12
Ladder Migration (mm)
Ladder Migration (mm)
Expon. (Ladder Migration (mm))
(X) 5300 (X) N/A N/A Inconclusive
It was concluded that the unknown plasmid was pBLU, this was found by looking into the data
above and reasoning that the closest one must be the correct one. The double digest had no
data that was relevant to my ladders, and as a consequence of such has been categorized as
inconclusive.
Sources and Footnotes:
1. Guruatma Ji Khalsa, 2010, Mama Ji's Molecular Kitchen. Internet:
http://askabiologist.asu.edu/plasmids.
2. Guruatma Ji Khalsa. 2010 "Mama Ji's Molecular Kitchen." Internet:
http://askabiologist.asu.edu/restriction-enzymes.
3. 2014. http://www.nature.com/scitable/definition/gel-electrophoresis-286
4. Vincze, T., Posfai, J. and Roberts, R.J.
NEBcutter: a program to cleave DNA with restriction enzymes
Nucleic Acids Res. 31: 3688-3691 (2003) http://tools.neb.com/NEBcutter2/
Você também pode gostar
- Hidden Figures: The American Dream and the Untold Story of the Black Women Mathematicians Who Helped Win the Space RaceNo EverandHidden Figures: The American Dream and the Untold Story of the Black Women Mathematicians Who Helped Win the Space RaceNota: 4 de 5 estrelas4/5 (895)
- The Subtle Art of Not Giving a F*ck: A Counterintuitive Approach to Living a Good LifeNo EverandThe Subtle Art of Not Giving a F*ck: A Counterintuitive Approach to Living a Good LifeNota: 4 de 5 estrelas4/5 (5794)
- The Yellow House: A Memoir (2019 National Book Award Winner)No EverandThe Yellow House: A Memoir (2019 National Book Award Winner)Nota: 4 de 5 estrelas4/5 (98)
- The Little Book of Hygge: Danish Secrets to Happy LivingNo EverandThe Little Book of Hygge: Danish Secrets to Happy LivingNota: 3.5 de 5 estrelas3.5/5 (400)
- Never Split the Difference: Negotiating As If Your Life Depended On ItNo EverandNever Split the Difference: Negotiating As If Your Life Depended On ItNota: 4.5 de 5 estrelas4.5/5 (838)
- Elon Musk: Tesla, SpaceX, and the Quest for a Fantastic FutureNo EverandElon Musk: Tesla, SpaceX, and the Quest for a Fantastic FutureNota: 4.5 de 5 estrelas4.5/5 (474)
- A Heartbreaking Work Of Staggering Genius: A Memoir Based on a True StoryNo EverandA Heartbreaking Work Of Staggering Genius: A Memoir Based on a True StoryNota: 3.5 de 5 estrelas3.5/5 (231)
- The Emperor of All Maladies: A Biography of CancerNo EverandThe Emperor of All Maladies: A Biography of CancerNota: 4.5 de 5 estrelas4.5/5 (271)
- Devil in the Grove: Thurgood Marshall, the Groveland Boys, and the Dawn of a New AmericaNo EverandDevil in the Grove: Thurgood Marshall, the Groveland Boys, and the Dawn of a New AmericaNota: 4.5 de 5 estrelas4.5/5 (266)
- The Hard Thing About Hard Things: Building a Business When There Are No Easy AnswersNo EverandThe Hard Thing About Hard Things: Building a Business When There Are No Easy AnswersNota: 4.5 de 5 estrelas4.5/5 (345)
- The World Is Flat 3.0: A Brief History of the Twenty-first CenturyNo EverandThe World Is Flat 3.0: A Brief History of the Twenty-first CenturyNota: 3.5 de 5 estrelas3.5/5 (2259)
- Team of Rivals: The Political Genius of Abraham LincolnNo EverandTeam of Rivals: The Political Genius of Abraham LincolnNota: 4.5 de 5 estrelas4.5/5 (234)
- The Unwinding: An Inner History of the New AmericaNo EverandThe Unwinding: An Inner History of the New AmericaNota: 4 de 5 estrelas4/5 (45)
- The Gifts of Imperfection: Let Go of Who You Think You're Supposed to Be and Embrace Who You AreNo EverandThe Gifts of Imperfection: Let Go of Who You Think You're Supposed to Be and Embrace Who You AreNota: 4 de 5 estrelas4/5 (1090)
- The Sympathizer: A Novel (Pulitzer Prize for Fiction)No EverandThe Sympathizer: A Novel (Pulitzer Prize for Fiction)Nota: 4.5 de 5 estrelas4.5/5 (121)
- 1 s2.0 S001346861530551X MainDocumento8 páginas1 s2.0 S001346861530551X Mainhenry.a.peraltaAinda não há avaliações
- Biochemical Aspects of CellDocumento39 páginasBiochemical Aspects of CellHaroon BadarAinda não há avaliações
- Medical Progress: Review ArticlesDocumento15 páginasMedical Progress: Review ArticlesFeri EspinosaSerranoAinda não há avaliações
- AdisDocumento15 páginasAdisadisu100% (1)
- BioinformaticsDocumento10 páginasBioinformaticsRamla FatimaAinda não há avaliações
- Primer3 Input Help: Start, Length Start Length Start, LengthDocumento2 páginasPrimer3 Input Help: Start, Length Start Length Start, LengthDiki Rizzal FauziAinda não há avaliações
- Brazilian Society For Food and Nutrition Position Statement - Nutrigenetic TestsDocumento12 páginasBrazilian Society For Food and Nutrition Position Statement - Nutrigenetic TestsDaniel Carvalho de AraújoAinda não há avaliações
- Cell Growth and DivisionDocumento21 páginasCell Growth and Divisionapi-319819727Ainda não há avaliações
- Revision Quiz MSDocumento30 páginasRevision Quiz MSPreya ShahAinda não há avaliações
- Amino Acids and PeptidesDocumento40 páginasAmino Acids and PeptidesDaniel LuchendoAinda não há avaliações
- Biomedical Importance Glycolysis Can Function Under Anaerobic ConditionsDocumento5 páginasBiomedical Importance Glycolysis Can Function Under Anaerobic ConditionsCatalina Denise Blaquera FloresAinda não há avaliações
- Overview of Post-Translational Modification AnalysisDocumento2 páginasOverview of Post-Translational Modification AnalysisSteven4654Ainda não há avaliações
- Untitled Document PDFDocumento4 páginasUntitled Document PDFBe different Be yourself online review ideas 2020Ainda não há avaliações
- Unit 9 Lipid MetabolismDocumento28 páginasUnit 9 Lipid MetabolismAthina100% (1)
- Chapter 29 - Amino Acids, Polypeptides and ProteinsDocumento120 páginasChapter 29 - Amino Acids, Polypeptides and Proteinssylv ackermanAinda não há avaliações
- Trusight One Sequencing Panel Reference Guide 15046431 03 PDFDocumento37 páginasTrusight One Sequencing Panel Reference Guide 15046431 03 PDFjoda23Ainda não há avaliações
- Cell Division and Inheritance PatternDocumento22 páginasCell Division and Inheritance PatternSheraz ChaudharyAinda não há avaliações
- The Birth and Death of Proteins: Some Key ConceptsDocumento2 páginasThe Birth and Death of Proteins: Some Key ConceptsLuke ShantiAinda não há avaliações
- DNA Paper Models ActivityDocumento8 páginasDNA Paper Models ActivityLouise Meara SeveroAinda não há avaliações
- GENBIO 1 Q1 Periodic TestDocumento6 páginasGENBIO 1 Q1 Periodic TestCelsa ChicoAinda não há avaliações
- Transposons Mobile DNADocumento6 páginasTransposons Mobile DNAkerkour-abd1523Ainda não há avaliações
- Lab Report Animal and Plant CellsDocumento5 páginasLab Report Animal and Plant CellsNicolas Gómez100% (1)
- Detection and Characterisation of Tobacco Leaf Curl Virus Isolates Infecting FCV Tobacco in IndiaDocumento5 páginasDetection and Characterisation of Tobacco Leaf Curl Virus Isolates Infecting FCV Tobacco in IndiaksaralaAinda não há avaliações
- Science Grade 9Documento41 páginasScience Grade 9Neil Janas100% (1)
- PCR CalculationsDocumento7 páginasPCR Calculationsmarwatsum100% (1)
- Biochemistry of Cheese RipeningDocumento19 páginasBiochemistry of Cheese RipeningKenesei GyörgyAinda não há avaliações
- 2014 JuneDocumento400 páginas2014 JunealiceAinda não há avaliações
- MLL4 Supplemental Figure 136155-JCI-RG-RV-3 - SD - 424851Documento15 páginasMLL4 Supplemental Figure 136155-JCI-RG-RV-3 - SD - 424851李博Ainda não há avaliações
- The Road To ModularityDocumento11 páginasThe Road To ModularityGiovanni DummondAinda não há avaliações
- Topic 1.3 Revision Practice (Ans)Documento3 páginasTopic 1.3 Revision Practice (Ans)Mahina WongAinda não há avaliações