Escolar Documentos
Profissional Documentos
Cultura Documentos
A NAC Domain Protein Interacts With Tomato Leaf Curl Virus Replication Accessory Protein and Enhances Viral Replication
Enviado por
kalyankpyTítulo original
Direitos autorais
Formatos disponíveis
Compartilhar este documento
Compartilhar ou incorporar documento
Você considera este documento útil?
Este conteúdo é inapropriado?
Denunciar este documentoDireitos autorais:
Formatos disponíveis
A NAC Domain Protein Interacts With Tomato Leaf Curl Virus Replication Accessory Protein and Enhances Viral Replication
Enviado por
kalyankpyDireitos autorais:
Formatos disponíveis
A NAC Domain Protein Interacts with Tomato leaf curl virus
Replication Accessory Protein and Enhances Viral
Replication
The Plant Cell, Vol. 17, 311–325, January 2005
Luke A. Selth, Satish C. Dogra, M. Saif Rasheed, Helen Healy,
John W. Randles and M. Ali Rezaiana,
Identification of Protein that interacts with REn
Yeast Two Hybrid Screening
• Tomato c-DNA library fused to B42 AD sequence (Trp)
• REn fused to the Lex A DNA BD (His)
• Reporters of interaction are growth on Leu- and GFP (Ura)
• Plasmid was rescued from colonies growing on Leu- with GFP
fluorescence.
• Retransformed into Yeast with Reporter and pLex-REn and
assessed to eliminate case of false positives.
• Identified a full length c-DNA for a protein of 301aa.
Nucleotide and amino acid sequences of
SlNAC1. The five subdomains (A to E)
comprising the NAC domain are shown in
empty boxes. A putative nuclear
localization signal is indicated by a bold
line under the sequence PRDRKYP. The
TAR-CM of the ATAF subgroup is also
boxed.
The predicted amino acid sequence of SlNAC1
(Figure 1A) and known NAC family proteins were
subjected to phylogenetic analysis. Multiple sequence
alignment of the proteins was conducted using
ClustalX (Thompson et al., 1997), and phylogenetic
analysis was performed by the neighborjoining
method (Saitou and Nei, 1987). A bootstrap analysis
of 1000 resampling replicates was conducted with
ClustalX. The rooted phylogenetic tree was displayed
using the NJPlot program included with ClustalX. The
gene names and references for other NACs are as
follows: A. thaliana, ATAF1 and
ATAF2 (Aida et al., 1997), AtNAC2 (Takada et al.,
2001), AtNAC3 (Takada et al., 2001), AtNAM (Duval
et al., 2002), CUC1 (Takada et al., 2001), CUC2
(Takada et al., 2001), CUC3 (Vroemen et al., 2003),
NAC1 (Xie et al., 2000), NAC2, NAP (Sablowski and
Meyerowitz, 1998), and TIP (Ren et al., 2000);
rice, OsNAC1 to OsNAC8 (Kikuchi et al., 2000);
petunia, NAM (Souer et al., 1996); tomato, SenU5
(John et al., 1997); potato, StNAC (Collinge and
Boller,
2001); and wheat, GRAB1 and GRAB2 (Xie et al.,
1999)
IDENTIFICATION OF INTERACTING REGIONS
Diagrammatic representation of REn (bait) and SlNAC1
proteins (prey) tested for interaction. The REn proteins
were expressed as LexA DNA BD fusions, and the
SlNAC1 proteins were expressed as B42 AD fusions.
The positions of three putative a-helices in REn are
indicated by closed boxes. In SlNAC1, the positions of
the NAC subdomains are shown in shaded boxes (A to
E), whereas the variable C terminus is denoted V.
The N-terminal region of REn is important for SlNAC1 binding.
Interaction was indicated by the ability of cells transformed
with bait, prey, and displayREPORTER plasmids to grow on
medium lacking Leu. As an additional indicator of interaction,
colonies were monitored for GFP expression by visualization
under UV light.
N- terminal region of REn is important for SlNAC1 binding
INTERACTION OF SLNAC1 WITH TGMV AND TLCV REn
Yeast two-hybrid assays testing the ability of SlNAC1 to
interact with REn of TLCV (REnTLCV) and TGMV
(REnTGMV). Yeast coexpressing proteins as indicated (top)
were grown on SD – His – Trp – uracil (Ura) medium
(bottom left), and interaction was tested by Leu prototrophy
and GFP expression on an inductive carbon source
(galactose and raffinose; bottom right). REn proteins were
fused to the LexA DNA BD, whereas SlNAC1 was fused to
the B42 AD. Negative controls included REnTLCV and
REnTGMV coexpressed with TLCV C2 fused to the AD, or
coexpressed with AD alone.
SlNAC1 Interacts with Both TLCV and TGMV REn.
SiNAC-1 : Role in Transcription
Regions of SlNAC1 able to activate transcription in yeast. The LexA:SlNAC1 fusion proteins are
represented diagrammatically at left, with the positions of the NAC subdomains shown in shaded
boxes (A to E) and the variable C terminus denoted V. The ability of LexA:SlNAC1 fusion proteins to
activate transcription in yeast is shown at right. Activities were assayed by measuring b-
galactosidase activity in total protein extracts from cells containing pLexA-SlNAC1 plasmids and
pSH18-34, which contains a lacZ reporter gene downstream of the LexA recognition site. Positive
control corresponds to yeast containing pSH18-34 and expressing a LexA fusion with the GAL4 AD.
Negative control corresponds to yeast containing pSH18- 34 and expressing LexA. Error bars
indicate the standard deviation for each sample.
C-terminal of SiNAC-1 has Trasncriptional Activation Domain
In Vitro Binding of SlNAC1 to TLCV REn
Purified 6X-His-tagged proteins were mixed with crude CBP-tagged protein mixtures, incubated with Ni-NTA,
and washed extensively to remove any unbound protein. Bound protein was resuspended in loading buffer,
resolved by SDS-PAGE, and analyzed by immunoblotting using anti-polyHis and anti-FLAG (CBP-tagged
proteins also contain a FLAG epitope) antibodies. Reactions were as follows: 6X-His-REn and CBPSlNAC1
(lane 5), 6X-His-REn and CBP-SlUPTG1 (lane 6), 6XHis-C2 and CBP-SlNAC1 (lane 7), and CBP-SlNAC1
alone (lane 8). Protein inputs for each reaction are shown: 6X-His-REn (lane 1), 6X-His-C2 (lane 2),
CBPSlNAC1 (lane 3), and CBP-SlUPTG1 (lane 4).
REn Interacts with SlNAC1 in Vitro
Localization of REn and SlNAC1
REn:GFP (top row) and SlNAC1:GFP (second
row), as well as GFP alone (bottom row), were
expressed in onion epidermal cells using the
CaMV 35S promoter after biolistic delivery of
vector DNA. A positive control for nuclear
localization, H2B:YFP, is also shown (third row).
Cells were analyzed for GFP and YFP
fluorescence (left column) by confocal
microscopy. Differential interference contrast
(DIC) images and merge images are shown in
the
middle and right columns, respectively. Nuclei in
merge images are indicated by arrows. Bar = 100
mm.
REn and SlNAC1 localize to the Nucleus of Onion Cells
Geminiviral Infection and Expression of SlNAC1
TYLCSV infection results in
an upregulation of SlNAC1
gene expression. (Northern
blot)
TLCV infection results in an upregulation A TLCV REn mutant cannot
of SlNAC1 gene expression. induce SlNAC1 gene
expression. RNA gel blot. The
presence of replicating TLCV
REn mutant was confirmed by
DNA gel blotting (middle). In
this blot, we also ran an
extract obtained from plants
infected with wild-type virus
(left, designated M); the ratio
of REn mutant:wild-type virus
total nucleic acid extracts is
Transient expression of REn is sufficient to induce SlNAC1 20:1. TLCV DNA species are
gene expression. Tomato leaves were infiltrated with A. marked RF (supercoiled
tumefaciens cells containing a replication-competent TLCV double-stranded replicative
1.1mer, p35S, or p35S expressing the TLCV genes C1, form) and SS (single
C2, and REn. stranded).
SlNAC1 is induced by TLCV Infection
Geminiviral Infection and Induction of SlNAC1
Tissue sections derived from mock-inoculated (A) and TLCV-infected ([B] to [G]) leaves of N. benthamiana were
hybridized with either fluorescein labeled ssRNA probe complementary to SlNAC1 ([A], [C], [D], [F], and [G]) or DIG-
labeled ssRNA probe complementary to TLCV ([A], [B], [D], [E], and [G]). (A) to (D) are cross sections and (E) to (G)
are longitudinal sections taken from the main leaf vein. Bar = 100 mm. Cell types present are indicated: E,
epidermal; M, mesophyll; P, phloem; and X, xylem.
Induction of SlNAC1 by TLCV Occurs Only in Infected Cells.
Effect of SlNAC1 Expression on TLCV DNA Accumulation
Analysis of SlNAC1 expression by p35S-SlNAC1 in
Expression of SlNAC1 enhances TLCV ssDNA
N. benthamiana leaf strips by semiquantitative RT-
accumulation in a transient replication assay.
PCR.
exposures.
SlNAC1 Expression Enhances TLCV ssDNA Accumulation
Probable Role of SlNAC1
• SlNAC1 might activate hosts cellular machinary for replication which
inturn might be utilised by virus for replication
• SlNAC1 might directly activate geminiviral replication like acidic
transcription factors ABF1
• SlNAC1 could positively modulate transcription of viral genes
Thank you……
Immunoblot analysis of yeast cells demonstrating that noninteracting
REn-LexA fusions and SlNAC1-B42 fusions are expressed at levels
similar to those of interacting fusion proteins. Total protein from yeast
cultures containing different REn and SlNAC1 fusion proteins was
extracted, fractionated on 4 to 20% SDS-polyacrylamide gels, and
immunoblotted with anti-LexA (to detect REn-LexA fusions) or antihemagglutinin
(HA) (to detect SlNAC1-B42 fusions).
Immunoblot analysis of yeast cells
demonstrating that nontransactivating
LexA:SlNAC1 fusions are expressed at
levels similar to those of
transactivating fusion proteins. Total
protein from yeast cultures containing
fusion proteins was extracted, fractionated
on SDS-polyacrylamide gels,
and immunoblotted with anti-LexA.
Você também pode gostar
- Inhibition of Virus DNA Replication by Artificial Zinc Finger ProteinsDocumento13 páginasInhibition of Virus DNA Replication by Artificial Zinc Finger ProteinskalyankpyAinda não há avaliações
- Application CSRDocumento1 páginaApplication CSRkalyankpyAinda não há avaliações
- Tomato Leaf Curl Kerala Virus (ToLCKeV) AC3 Protein Forms A Higher Order Oligomer and Enhances ATPase Activity of Replication Initiator Protein (Rep/AC1)Documento22 páginasTomato Leaf Curl Kerala Virus (ToLCKeV) AC3 Protein Forms A Higher Order Oligomer and Enhances ATPase Activity of Replication Initiator Protein (Rep/AC1)kalyankpyAinda não há avaliações
- Full Page PhotoDocumento28 páginasFull Page PhotokalyankpyAinda não há avaliações
- Principles of MicroRNA-Target RecognitionDocumento22 páginasPrinciples of MicroRNA-Target RecognitionkalyankpyAinda não há avaliações
- Survey: Are You Registered For PHDDocumento5 páginasSurvey: Are You Registered For PHDkalyankpyAinda não há avaliações
- The Nanovirus-Encoded Clink Protein Affects Plant Cell Cycle Regulation Through Interaction With The Retinoblastoma-Related ProteinDocumento11 páginasThe Nanovirus-Encoded Clink Protein Affects Plant Cell Cycle Regulation Through Interaction With The Retinoblastoma-Related ProteinkalyankpyAinda não há avaliações
- Characterization of The piRNA Complex From Rat TestesDocumento8 páginasCharacterization of The piRNA Complex From Rat TesteskalyankpyAinda não há avaliações
- Expression of A Viral Polymerase-Bound Host Factor Turns Human Cell Lines Permissive To A Plant - and Insect-Infecting VirusDocumento10 páginasExpression of A Viral Polymerase-Bound Host Factor Turns Human Cell Lines Permissive To A Plant - and Insect-Infecting ViruskalyankpyAinda não há avaliações
- BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program For Windows 95/98/NTDocumento14 páginasBioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program For Windows 95/98/NTkalyankpyAinda não há avaliações
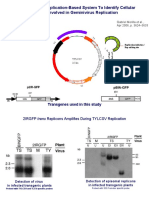
- A Versatile Transreplication-Based System To Identify Cellular Proteins Involved in Geminivirus ReplicationDocumento6 páginasA Versatile Transreplication-Based System To Identify Cellular Proteins Involved in Geminivirus ReplicationkalyankpyAinda não há avaliações
- PHD Viva - 5nov09Documento65 páginasPHD Viva - 5nov09kalyankpyAinda não há avaliações
- The Mechanism of DNA Replication Primer Synthesis by RNA PolymeraseDocumento9 páginasThe Mechanism of DNA Replication Primer Synthesis by RNA PolymerasekalyankpyAinda não há avaliações
- Peptide Aptamers That Bind To A Geminivirus Replication Protein Interfere With Viral Replication in Plant CellsDocumento28 páginasPeptide Aptamers That Bind To A Geminivirus Replication Protein Interfere With Viral Replication in Plant CellskalyankpyAinda não há avaliações
- MSC ProjectDocumento29 páginasMSC ProjectkalyankpyAinda não há avaliações
- Identification of Sequence Elements Regulating Promoter Activity and Replication of a Monopartite Begomovirus-Associated DNA β satelliteDocumento10 páginasIdentification of Sequence Elements Regulating Promoter Activity and Replication of a Monopartite Begomovirus-Associated DNA β satellitekalyankpyAinda não há avaliações
- Regulation of Replication Fork Progression Through Histone Supply and DemandDocumento24 páginasRegulation of Replication Fork Progression Through Histone Supply and DemandkalyankpyAinda não há avaliações
- Vector MapsDocumento14 páginasVector MapskalyankpyAinda não há avaliações
- Next Generation Sequencing PresentationDocumento28 páginasNext Generation Sequencing PresentationkalyankpyAinda não há avaliações
- Paper RNAi in Yeast LS 3oct09Documento24 páginasPaper RNAi in Yeast LS 3oct09kalyankpyAinda não há avaliações
- PHDDocumento1 páginaPHDkalyankpyAinda não há avaliações
- National Symposium PosterDocumento1 páginaNational Symposium PosterkalyankpyAinda não há avaliações
- GateDocumento1 páginaGatekalyankpyAinda não há avaliações
- Role of AC3 On Geminiviral Replication: A Journey Through PastDocumento25 páginasRole of AC3 On Geminiviral Replication: A Journey Through PastkalyankpyAinda não há avaliações
- Complete Thesis 23jul09Documento130 páginasComplete Thesis 23jul09kalyankpyAinda não há avaliações
- Mechanism of Rolling Circle Replication in Geminiviruses: Role of Replication Enhancer (Ren/Al3)Documento18 páginasMechanism of Rolling Circle Replication in Geminiviruses: Role of Replication Enhancer (Ren/Al3)kalyankpy100% (1)
- PHD Course Work DrKhannaDocumento63 páginasPHD Course Work DrKhannakalyankpyAinda não há avaliações
- MSC PaperpptDocumento16 páginasMSC PaperpptkalyankpyAinda não há avaliações
- MSC PaperpptDocumento16 páginasMSC PaperpptkalyankpyAinda não há avaliações
- The Subtle Art of Not Giving a F*ck: A Counterintuitive Approach to Living a Good LifeNo EverandThe Subtle Art of Not Giving a F*ck: A Counterintuitive Approach to Living a Good LifeNota: 4 de 5 estrelas4/5 (5794)
- The Yellow House: A Memoir (2019 National Book Award Winner)No EverandThe Yellow House: A Memoir (2019 National Book Award Winner)Nota: 4 de 5 estrelas4/5 (98)
- Hidden Figures: The American Dream and the Untold Story of the Black Women Mathematicians Who Helped Win the Space RaceNo EverandHidden Figures: The American Dream and the Untold Story of the Black Women Mathematicians Who Helped Win the Space RaceNota: 4 de 5 estrelas4/5 (895)
- The Hard Thing About Hard Things: Building a Business When There Are No Easy AnswersNo EverandThe Hard Thing About Hard Things: Building a Business When There Are No Easy AnswersNota: 4.5 de 5 estrelas4.5/5 (344)
- The Little Book of Hygge: Danish Secrets to Happy LivingNo EverandThe Little Book of Hygge: Danish Secrets to Happy LivingNota: 3.5 de 5 estrelas3.5/5 (399)
- The Emperor of All Maladies: A Biography of CancerNo EverandThe Emperor of All Maladies: A Biography of CancerNota: 4.5 de 5 estrelas4.5/5 (271)
- Devil in the Grove: Thurgood Marshall, the Groveland Boys, and the Dawn of a New AmericaNo EverandDevil in the Grove: Thurgood Marshall, the Groveland Boys, and the Dawn of a New AmericaNota: 4.5 de 5 estrelas4.5/5 (266)
- Never Split the Difference: Negotiating As If Your Life Depended On ItNo EverandNever Split the Difference: Negotiating As If Your Life Depended On ItNota: 4.5 de 5 estrelas4.5/5 (838)
- A Heartbreaking Work Of Staggering Genius: A Memoir Based on a True StoryNo EverandA Heartbreaking Work Of Staggering Genius: A Memoir Based on a True StoryNota: 3.5 de 5 estrelas3.5/5 (231)
- Elon Musk: Tesla, SpaceX, and the Quest for a Fantastic FutureNo EverandElon Musk: Tesla, SpaceX, and the Quest for a Fantastic FutureNota: 4.5 de 5 estrelas4.5/5 (474)
- Team of Rivals: The Political Genius of Abraham LincolnNo EverandTeam of Rivals: The Political Genius of Abraham LincolnNota: 4.5 de 5 estrelas4.5/5 (234)
- The World Is Flat 3.0: A Brief History of the Twenty-first CenturyNo EverandThe World Is Flat 3.0: A Brief History of the Twenty-first CenturyNota: 3.5 de 5 estrelas3.5/5 (2259)
- The Unwinding: An Inner History of the New AmericaNo EverandThe Unwinding: An Inner History of the New AmericaNota: 4 de 5 estrelas4/5 (45)
- The Gifts of Imperfection: Let Go of Who You Think You're Supposed to Be and Embrace Who You AreNo EverandThe Gifts of Imperfection: Let Go of Who You Think You're Supposed to Be and Embrace Who You AreNota: 4 de 5 estrelas4/5 (1090)
- The Sympathizer: A Novel (Pulitzer Prize for Fiction)No EverandThe Sympathizer: A Novel (Pulitzer Prize for Fiction)Nota: 4.5 de 5 estrelas4.5/5 (120)
- Basf Sharontan 101216215349 Phpapp01Documento29 páginasBasf Sharontan 101216215349 Phpapp01Soyeb HassanAinda não há avaliações
- Assignment 1 - CarbohydratesDocumento4 páginasAssignment 1 - CarbohydratesJay Ann BernalesAinda não há avaliações
- 3D Optical and Device Simulation of Surface Plasmonic Effects On Organic Solar Cells Using Silver Nano PrismsDocumento5 páginas3D Optical and Device Simulation of Surface Plasmonic Effects On Organic Solar Cells Using Silver Nano PrismsSakshiKoulAinda não há avaliações
- The Mineral Shadows CollectionDocumento6 páginasThe Mineral Shadows CollectionalexkardarAinda não há avaliações
- ME324 Jeopardy2Documento22 páginasME324 Jeopardy2Catherine AndersonAinda não há avaliações
- Fatigue Analysis W - CIIDocumento10 páginasFatigue Analysis W - CIInaeandAinda não há avaliações
- Topical, Gastrointestinal, NI, Radio-, MiscDocumento113 páginasTopical, Gastrointestinal, NI, Radio-, MiscArk Olfato ParojinogAinda não há avaliações
- Application of Essential Oils in Food SystemsDocumento116 páginasApplication of Essential Oils in Food SystemsMădălina Ștefan100% (1)
- Biomass FinalDocumento12 páginasBiomass FinalAbu ikhtiarAinda não há avaliações
- 2Documento26 páginas2Aigerim TurlanovaAinda não há avaliações
- (De Dios) Science ProjectDocumento22 páginas(De Dios) Science ProjectRoucyzle Ynnah AcidoAinda não há avaliações
- WellLock Resin Rigless Micro Channel Remediation H011331Documento1 páginaWellLock Resin Rigless Micro Channel Remediation H011331bagus918Ainda não há avaliações
- Dead Molecules and The Live Organism: Moh. Dliyauddin (176090100111019) Rubiyatna Sakaroni (176090100111006)Documento14 páginasDead Molecules and The Live Organism: Moh. Dliyauddin (176090100111019) Rubiyatna Sakaroni (176090100111006)Muhammad DliyauddinAinda não há avaliações
- Shell Structures PDFDocumento21 páginasShell Structures PDFDivya VarshneyAinda não há avaliações
- A Helmholtz Free Energy Formulation of The Thermodynamic Properties of The Mixture (Water + Ammonia)Documento35 páginasA Helmholtz Free Energy Formulation of The Thermodynamic Properties of The Mixture (Water + Ammonia)Marcelo SilvosaAinda não há avaliações
- Van Urk Indole TLC Test PDFDocumento11 páginasVan Urk Indole TLC Test PDFabazaba151Ainda não há avaliações
- Well Classification SystemDocumento2 páginasWell Classification SystemMuhammad Naufal Nazhib KhanAinda não há avaliações
- High Performance Fiber Reinforced ConcreteDocumento11 páginasHigh Performance Fiber Reinforced ConcreteZaireen AzmeeAinda não há avaliações
- Welding F22 To F91 (09MAR2018)Documento2 páginasWelding F22 To F91 (09MAR2018)Juan Shuna100% (2)
- Classes of CompoundsDocumento12 páginasClasses of CompoundsReynaldo VirtucioAinda não há avaliações
- Catalogue CSM PDFDocumento4 páginasCatalogue CSM PDFFran IgledominguezAinda não há avaliações
- D BlockDocumento17 páginasD Block145556Ainda não há avaliações
- Revision Notes - Basic Nuclear PropertiesDocumento10 páginasRevision Notes - Basic Nuclear PropertiesPankaj BiswasAinda não há avaliações
- Introduction To Flip ChipDocumento58 páginasIntroduction To Flip ChipLakshman Yandapalli100% (1)
- Brenntag Tofa Data Sheet: BFA 101 - Tall Oil Fatty AcidDocumento1 páginaBrenntag Tofa Data Sheet: BFA 101 - Tall Oil Fatty AcidniteshacharyaAinda não há avaliações
- Nirosta 4104: Krupp EdelstahlprofileDocumento2 páginasNirosta 4104: Krupp EdelstahlprofileLuis MayorgaAinda não há avaliações
- Glucose o ToluidineDocumento21 páginasGlucose o ToluidinetorokpeterAinda não há avaliações
- Fuel Gas Conditioning-EmailDocumento2 páginasFuel Gas Conditioning-Emailkelburn50% (2)
- Safety Data Sheet: Section 1 - Identification of The Substance and CompanyDocumento6 páginasSafety Data Sheet: Section 1 - Identification of The Substance and CompanyleonardovegaAinda não há avaliações
- FY 2018 Self Identified Generic Drug Facilities Sites and OrganizationsDocumento175 páginasFY 2018 Self Identified Generic Drug Facilities Sites and OrganizationsDavid Paul HensonAinda não há avaliações